A decade of improvements on the reference green alga genome
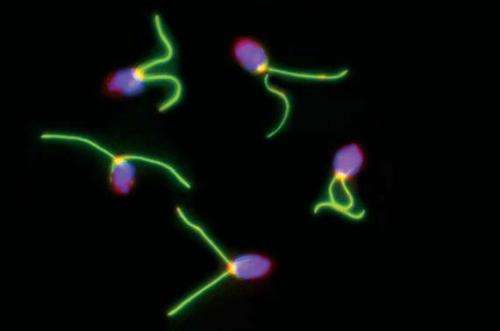
The high-quality genome sequence of the tiny single-celled alga Chlamydomonas reinhardtii has proved useful for researchers studying photosynthesis and cell motility.
In the decade since researchers initiated plans to sequence and now annotate the Chlamydomonas genome, constant improvements to the genome assembly have been made.
Estimated to be just one-fiftieth the size of a grain of salt, the green alga Chlamydomonas reinhardtii (referred to by many as simply "Chlamy") interests researchers because its evolutionary place in the timeline when plants and animals diverged means it has features found in animals but lost in land plants. For the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science user facility, however, the alga's photosynthetic capabilities are one of the drivers behind sequencing its genome. Understanding these pathways could lead to improving plant biomass yield for biofuels production.
In a perspective published online June 17, 2014 in Trends in Plant Science, a team led by DOE JGI researchers review the work done on the Chlamydomonas genome in the past decade and look at future plans. Since the draft algal genome was published in Science seven years ago, DOE JGI researchers have worked on improving the sequence data available. For example, in 2012, the genome assembly made publicly available to the research community was already in its 5th iteration and utilized sequence generated through two different sequencing technologies. More importantly, gene model predictions over the years have been substantially improved, in part through experiments that provided gene expression data.
"Owing to the high-quality genome sequence and the substantial amount of expression data available," the team wrote, "as well as the functional annotation efforts of the community, gene models in the DOE JGI flagship genome of Chlamydomonas represent the most highly curated genomic data for any alga." All of the omics data about the alga, including the latest gene model (version 5.5) to be released later this year, is publicly available on Phytozome, the DOE JGI's plant genomics portal.
The plethora of information available has also led the DOE JGI research team and their collaborators to develop guidelines for naming genes and developing gene symbols. "If future gene naming follows the policy outlined," they noted, "this will help maximize the benefits that the Chlamydomonas community derives from its genome project, particularly as refinements and developments continue into the future."
More information: Blaby IK et al. "The Chlamydomonas genome project: a decade on." Trends Plant Sci. 2014 Jun 17. pii: S1360-1385(14)00143-5. DOI: 10.1016/j.tplants.2014.05.008. [Epub ahead of print]
Journal information: Trends in Plant Science
Provided by US Department of Energy




















