This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
PMAT: Researchers develop a new tool for efficient assembly of plant mitochondrial genomes
Plant mitochondrial genomes (mitogenomes) are crucial for understanding nucleocytoplasmic interactions, plant evolution, and breeding of cytoplasmic male sterile lines. However, their complete assembly is challenging due to frequent recombination events and horizontal gene transfers.
Traditional methods using Illumina, PacBio, and Nanopore sequencing data often result in poor assembly completeness, low sequencing accuracy, and high costs, limiting their applicability. Based on these challenges, there is a need for a more efficient and accurate assembly method to conduct in-depth research on plant mitogenomes.
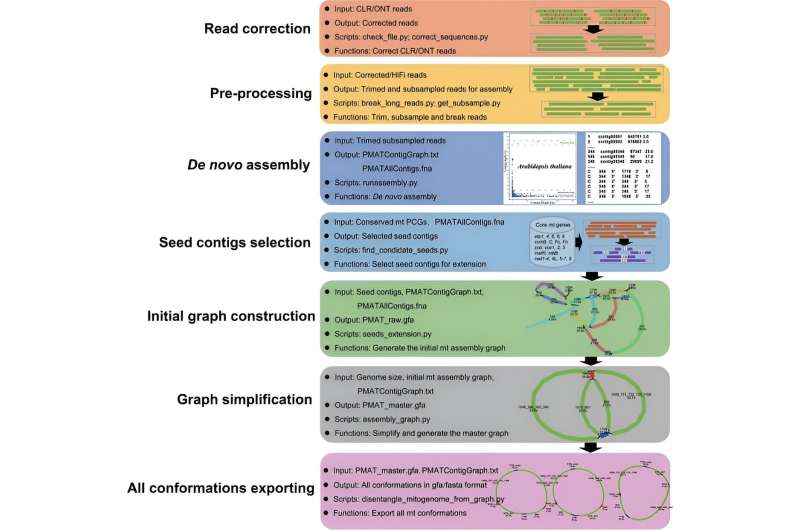
Researchers from Nanjing Forestry University, in collaboration with the Beijing Academy of Agriculture and Forestry Sciences and the Chinese Academy of Agricultural Sciences, have developed an efficient assembly toolkit (PMAT) for de novo assembly of plant mitogenomes using low-coverage HiFi sequencing data.
The study was published in the journal Horticulture Research on January 26 2024, showcasing a significant leap in genomics research.
PMAT addresses the limitations of traditional mitochondrial genome assembly methods by utilizing highly accurate long-read HiFi sequencing data. This allows for the spanning of most repeats and the generation of complete and accurate mitochondrial genome sequences.
The toolkit includes two modes: "autoMito" and "graphBuild." The "autoMito" mode provides a one-step assembly process, while the "graphBuild" mode allows for manual selection of appropriate seeds for assembly, ensuring flexibility and user control.
The researchers successfully assembled the mitogenomes of 13 plant species, including eudicots, monocots, and gymnosperms. For example, the Arabidopsis thaliana mitochondrial genome was reassembled into a typical single circular chromosome with a length of 367,810 base pairs, showing only minor differences from the published reference genome.
In addition, PMAT required minimal sequencing data to achieve complete assemblies, making it a cost-effective solution for large-scale genomic studies.
Dr. Zhiqiang Wu, one of the leading researchers, commented, "PMAT represents a significant advancement in the field of plant genomics. By overcoming the challenges of traditional assembly methods, PMAT provides a comprehensive and accurate view of plant mitogenomes, facilitating deeper insights into plant evolution and breeding."
The development of PMAT significantly impacts plant genomic research and breeding. By offering a reliable method for assembling plant mitogenomes, PMAT accelerates studies on genome variation and its effects on plant traits. This enhances breeding efforts to improve crop resilience, yield, and quality.
Additionally, capturing multiple mitochondrial conformations opens new research avenues into the evolutionary dynamics of plant genomes.
More information: Changwei Bi et al, PMAT: an efficient plant mitogenome assembly toolkit using low-coverage HiFi sequencing data, Horticulture Research (2024). DOI: 10.1093/hr/uhae023
Journal information: Horticulture Research
Provided by TranSpread