This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A small factor makes a big impact on genome editing

Through years of engineering gene-editing systems, researchers have developed a suite of tools that enable the modification of genomes in living cells, akin to "genome surgery." These tools, including ones based on a natural system known as CRISPR/Cas9, offer enormous potential for addressing unmet clinical needs, underscored by the recent FDA approval of the first CRISPR/Cas9-based therapy.
A relatively new approach called "prime editing" enables gene-editing with exceptional accuracy and high versatility, but has a critical tradeoff: variable and often low efficiency of edit installation. In other words, while prime edits can be made with high precision and few unwanted byproducts, the approach also often fails to make those edits at reasonable frequencies.
In a paper that appeared in print in the journal Nature on April 18, 2024, Princeton scientists Jun Yan and Britt Adamson, along with several colleagues, describe a more efficient prime editor.
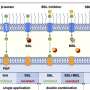
Prime editing systems minimally consist of two components: a modified version of the protein element of CRISPR/Cas9 and a ribonucleic acid (RNA) molecule called a pegRNA. These components work together in several coordinated steps: First, the pegRNA binds the protein and guides the resulting complex to a desired location in the genome.
There, the protein nicks the DNA and, using a template sequence encoded on the pegRNA, "reverse transcribes" an edit into the genome nearby. In this way, prime editors "write" exact sequences into targeted DNA.
"Prime editing is such an incredibly powerful genome editing tool because it gives us more control over exactly how genomic sequences are changed," Adamson said.
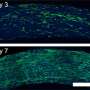
At the outset of their study, Adamson and Yan, a graduate student in Adamson's research group and the Department of Molecular Biology, reasoned that unknown cellular processes may aid or hinder prime editing. To identify such processes, Yan laid out a conceptually simple plan: First, he would engineer a cell line that would emit green fluorescence when certain prime edits were installed. Then, he would systematically block expression of proteins normally expressed within those cells and measure editing-induced fluorescence to determine which of those proteins impact prime editing.
By executing this plan, the team identified 36 cellular determinants of prime editing, only one of which—the small RNA-binding protein La—promoted editing.
"Although promoting prime editing is obviously not a normal function of the La protein, our experiments showed that it can strongly facilitate the process," Yan said.
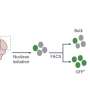
Within cells, La is known to bind specific sequences often found at the ends of nascent small RNA molecules and it protects those RNAs from degradation. The Princeton team recognized right away that the pegRNAs deployed in Yan's first experiments likely contained those exact sequences, called polyuridine tracts, as they are a typical but often overlooked byproduct of pegRNA expression in cells. Subsequent experiments suggested that such pegRNAs inadvertently harness La's end-binding activity for protection and to promote prime editing.
Motivated by their results, the team asked if fusing the part of La that binds polyuridine tracts to a standard prime editing protein could boost prime editing efficiencies. They were thrilled to find that the resulting protein, which they call PE7, substantially enhanced intended prime editing efficiencies across conditions and, when using some prime editing systems, left the frequencies of unwanted byproducts very low.
Their results quickly drew the attention of colleagues interested in using prime editing in primary human cells, including Daniel Bauer at Boston Children's Hospital and Harvard Medical School and Alexander Marson at the University of California, San Francisco. Together with scientists from these labs, the team of researchers went on to demonstrate that PE7 can also enhance prime editing efficiencies in therapeutically relevant cell types, offering expanded promise for future clinical applications.
"This work is a beautiful example of how deeply probing the inner workings of cells can lead to unexpected insights that may yield near-term biomedical impact," Bauer noted.
More information: Jun Yan et al, Improving prime editing with an endogenous small RNA-binding protein, Nature (2024). DOI: 10.1038/s41586-024-07259-6
Journal information: Nature
Provided by Princeton University