Watching detox enzymes work could lead to better medicines
Medicines that are easier on your liver, as well as on the environment, could one day result from a new experimental technique developed by University of Michigan researchers.
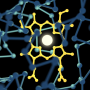
The researchers figured out how to gently trap a class of key enzymes called Cytochrome P450s, which serve as cleansing proteins in the liver and are vital to drug digestion. Lead researcher Ayyalusamy Ramamoorthy describes the class as "Mother Nature's blowtorch," because of its role in oxidizing, or burning, potentially toxic compounds.
Using the new trap procedure, scientists can watch how the enzyme functions in a 3D environment that mimics its natural surroundings—and at atomistic resolution. Current techniques can't deliver that.
Why watch Cytochrome P450s work?
"This family of enzymes serves as the detoxifier in our liver cells," said Ramamoorthy, who is the Robert W. Parry Collegiate Professor of Chemistry and Biophysics in the College of Literature, Science, and the Arts. "It provides a cleaning mechanism. Understanding its function is crucial for a healthy society."
Cytochrome P450s are ubiquitous in living things. In humans, this family of enzymes is responsible for breaking down 70 percent of the pharmaceutical drugs on the current market.
"Now we can really study these proteins in native condition. We can look at their motion instead of just their structure," Ramamoorthy said. "This could enable us to see the exact pathway that the drugs take as they make their way into the catalytic domain of the enzyme, where they get broken down. Eventually this could help us to know how to design drugs that the body can absorb more efficiently."
Our bodies can't currently squeeze all the active ingredients out of medications, and so today a lot of those compounds go to waste—literally. We excrete them in urine and they end up in waterways where they can disrupt ecosystems and could one day be a health concern for humans.
To create their new trapping system, the U-M researchers made snippets of artificial cell membrane—oily, flat nanoscale disks that the enzymes would bind to when they mingled with the disks in a solution.
Previously, when scientists wanted to study this enzyme, they'd have to snip off its membrane-binding portion and crystalize what was left—freeze it in stopped motion. That wasn't giving the complete picture.
"If you want to understand the function of a biological molecule, you have to understand its structure and its dynamics. This lets us do that," Ramamoorthy said, adding that their membranes are stable for several weeks to months.
The scientists have already uncovered pertinent information about the workings of this enzyme family. They've observed how it coordinates with a partnering protein to exchange electrons, kick starting the electron transfer chain that eventually metabolizes drugs. The coordination with this partnering protein happens in what Ramamoorthy calls "microsecond claps."
The researchers used a non-invasive imaging technique called nuclear magnetic resonance, which is similar to MRI, to reveal the structure of the complexes formed between cytochrome P450 and its protein partners in the liver cell membrane.
"It is remarkable that nuclear magnetic resonance has provided an atomistic map of the areas of contact between these proteins," Ramamoorthy said.
The findings, he adds, have the potential to illuminate for researchers how drug compounds affect the speed of that microsecond clapping action, which is directly tied to how efficiently drugs are metabolized. The quicker the clapping, the more molecules "Mother Nature's blowtorch" can turn its heat on.
A paper on the findings titled "Reconstitution of Cytb5–CytP450 Complex in Nanodiscs for Structural Studies by NMR" is newly published online in the journal Angewandte Chemie.
More information: Meng Zhang et al. Reconstitution of the Cyt -CytP450 Complex in Nanodiscs for Structural Studies using NMR Spectroscopy , Angewandte Chemie International Edition (2016). DOI: 10.1002/anie.201600073
Journal information: Angewandte Chemie , Angewandte Chemie International Edition
Provided by University of Michigan