Unpacking the mysteries of bacterial cell cycle regulation
As part of their long-term investigation of regulatory factors in the bacterial cell cycle, molecular biologists at the University of Massachusetts Amherst now report finding a surprising new role for one factor, CpdR, an adaptor that helps to regulate selective protein destruction, the main control mechanism of cell cycle progression in bacteria, at specific times.
Joanne Lau, a microbiology doctoral student, and her advisor Peter Chien in the biochemistry and molecular biology department at UMass Amherst, report how two molecular pathways, protein degradation and phosphorylation, work together to ensure normal bacterial growth, in a recent early online edition of Molecular Cell. Chien says that because controlled protein degradation is critical for bacterial virulence, "this work introduces new pathways for us to target in the discovery of sorely needed new antibiotics."
In bacteria, the cell cycle is very tightly controlled by protein-digesting enzymes called proteases that selectively destroy other proteins, called substrates, at appropriate times while the cell is undergoing growth and division. At the same time, another process called phosphorylation chemically modifies different proteins to regulate their activity in a cell-cycle-dependent way, Chien explains.
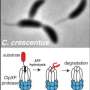
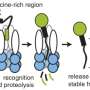
Although the energy-dependent proteases that carry out most protein degradation can directly recognize some substrates, biological regulation often requires additional factors known as adaptors to be present to "tune" substrate selection more precisely. In the bacterium Caulobacter crescentus studied in the Chien lab, one of these adaptors, a small protein called CpdR, is specifically phosphorylated at different times in the cell cycle. Previous studies had shown that the timing of this phosphorylation correlated with the degradation of many proteins by a protease called ClpXP.
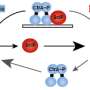
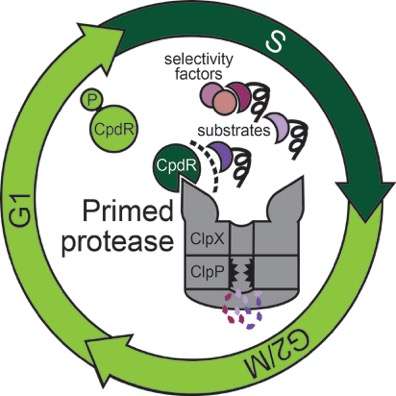
Chien says, "Before this work, we thought most adaptors were binding to the protease and substrate at the same time, effectively leashing them together to force them to interact. Surprisingly, Joanne found that CpdR bound principally to the ClpXP protease, but didn't seem to bind the substrate well at all. Instead, CpdR binding to the ClpXP protease prepared, or primed, the protease for engaging substrates. This primed protease was now also able to recruit additional adaptors that could deliver even more protease substrates."
He adds, "By not specifically interacting with any single substrate, this new mechanism of protease priming allows for surprisingly broad recognition of both substrates and additional regulators. This mechanism lets the cell control multiple pathways with a single regulator, which is useful when bacteria have to respond rapidly, such as during the stress they undergo when treated with antibiotics. However, this could also lead to unwanted degradation of off-target proteins. Understanding this balance of specificity and broad recognition is an outstanding question."
Chien and Lau say the "fascinating mystery" at the heart of this work has been to figure out how the cell cycle, made up of the many events that happen when cells divide, are so precisely coordinated by CpdR and other factors. The researchers say their new understanding of how adaptors work could answer the question of how a single regulator like CpdR could globally control degradation by a protease, ClpXP.
More information: Molecular Cell, www.cell.com/molecular-cell/ab … 1097-2765(15)00350-0
Journal information: Molecular Cell
Provided by University of Massachusetts Amherst