Some genes 'foreign' in origin and not from our ancestors
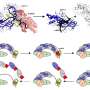
Many animals, including humans, acquired essential 'foreign' genes from microorganisms co-habiting their environment in ancient times, according to research published in the open access journal Genome Biology. The study challenges conventional views that animal evolution relies solely on genes passed down through ancestral lines, suggesting that, at least in some lineages, the process is still ongoing.
The transfer of genes between organisms living in the same environment is known as horizontal gene transfer (HGT). It is well known in single-celled organisms and thought to be an important process that explains how quickly bacteria evolve, for example, resistance to antibiotics.
HGT is thought to play an important role in the evolution of some animals, including nematode worms which have acquired genes from microorganisms and plants, and some beetles that gained bacterial genes to produce enzymes for digesting coffee berries. However, the idea that HGT occurs in more complex animals, such as humans, rather than them solely gaining genes directly from ancestors, has been widely debated and contested.
Lead author Alastair Crisp from the University of Cambridge, UK, said: "This is the first study to show how widely horizontal gene transfer (HGT) occurs in animals, including humans, giving rise to tens or hundreds of active 'foreign' genes. Surprisingly, far from being a rare occurrence, it appears that HGT has contributed to the evolution of many, perhaps all, animals and that the process is ongoing, meaning that we may need to re-evaluate how we think about evolution."
The researchers studied the genomes of 12 species of Drosophila or fruit fly, four species of nematode worm, and 10 species of primate, including humans. They calculated how well each of their genes aligns to similar genes in other species to estimate how likely they were to be foreign in origin. By comparing with other groups of species, they were able to estimate how long ago the genes were likely to have been acquired.
A number of genes, including the ABO blood group gene, were confirmed as having been acquired by vertebrates through HGT. The majority of the other genes were related to enzymes involved in metabolism.
In humans, they confirmed 17 previously-reported genes acquired from HGT, and identified 128 additional foreign genes in the human genome that have not previously been reported.
Some of those genes were involved in lipid metabolism, including the breakdown of fatty acids and the formation of glycolipids. Others were involved in immune responses, including the inflammatory response, immune cell signalling, and antimicrobial responses, while further gene categories include amino-acid metabolism, protein modification and antioxidant activities.
The team were able to identify the likely class of organisms the transferred genes came from. Bacteria and protists, another class of microorganisms, were the most common donors in all species studied. They also identified HGT from viruses, which was responsible for up to 50 more foreign genes in primates.
Some genes were identified as having originated from fungi. This explains why some previous studies, which only focused on bacteria as the source of HGT, originally rejected the idea that these genes were 'foreign' in origin.
The majority of HGT in primates was found to be ancient, occurring sometime between the common ancestor of Chordata and the common ancestor of the primates.
The authors say that their analysis probably underestimates the true extent of HGT in animals and that direct HGT between complex multicellular organisms is also plausible, and already known in some host-parasite relationships.
The study also has potential impacts on genome sequencing more generally. Genome projects frequently remove bacterial sequences from results on the assumption that they are contamination. While screening for contamination is necessary, the potential for bacterial sequences being a genuine part of an animal's genome originating from HGT should not be ignored, say the authors.
More information: Alastair Crisp, Chiara Boschetti, Malcolm Perry, Alan Tunnacliffe and Gos Micklem, Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes, Genome Biology 2015. DOI: 10.1186/s13059-015-0607-3
Journal information: Genome Biology
Provided by BioMed Central