April 25, 2014 report
Researchers use cryo-electron microscopy to learn how DNA wraps tightly around nucleosomes
(Phys.org) —A team of researchers working at the Chinese Academy of Sciences, in Beijing, has used cryo-electron microscopy to reveal how it is that DNA wraps so tightly around nuclesomes. In their paper published in the journal Science, the team describes how they managed to identify the path of linker DNA. Andrew Travers offers a Perspective piece in the same issue explaining the team's findings and what it might mean for DNA research in the future.
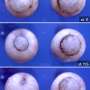
DNA in eukaryote cells (those with a nucleus surrounded by a membrane) comes packaged inside of what are known as nucleosomes, which are materials made up primarily of histone proteins—together they form chromatin. Prior research has shown that nucleosomes exist (due to folding) as coiled structured helix shapes, approximately 30nm in diameter. Inside each helix is a "linker" histone—a fiber that runs the length of the helix. Up until now, the molecular makeup of the linker has been a bit of a mystery, with competing researchers suggesting different ideas. In this new effort, the researchers in China appear to have settled the debate using cryo-electron microscopy, a form of transition electron microscopy where the material being studied is kept at cryogenic temperatures. Examination by the team revealed the linker histone fiber to be constructed from arrays of 12 nucleosomes.
The arrayed nucleosomes constitute the longest segment of chromatin observed to date, and help explain how it is that genomes can be packaged, yet remain easily accessible. The researchers found that the linker histone (type H1) binds nucleosomes together into structures known as tetranucleosomal units—they in turn form double-helical structures similar to the DNA that they serve to package. The team notes that the binding that occurs results in stacking, which is how the double-helix is able to form.
The findings by the research team point out the important role H1 linker histones play in stacking nucleosomes, which, they note, serves to make chromatin compact. They also note that nucleosomes can fold into other helical structures under different circumstances, which opens the door to other research efforts in the future. Also as Travers points out, their results lead to other questions, such as whether the structures they've uncovered correspond to the fiber in its natural state or if there are others. There's also the question of what happens when the helix is unwound, does it cause changes to internucleosomal interactions? Finding answers to such questions will likely be the focus of the research team in China as well as others working in the field for many years to come.
More information: Cryo-EM Study of the Chromatin Fiber Reveals a Double Helix Twisted by Tetranucleosomal Units, Science 25 April 2014: Vol. 344 no. 6182 pp. 376-380. DOI: 10.1126/science.1251413
ABSTRACT
The hierarchical packaging of eukaryotic chromatin plays a central role in transcriptional regulation and other DNA-related biological processes. Here, we report the 11-angstrom–resolution cryogenic electron microscopy (cryo-EM) structures of 30-nanometer chromatin fibers reconstituted in the presence of linker histone H1 and with different nucleosome repeat lengths. The structures show a histone H1-dependent left-handed twist of the repeating tetranucleosomal structural units, within which the four nucleosomes zigzag back and forth with a straight linker DNA. The asymmetric binding and the location of histone H1 in chromatin play a role in the formation of the 30-nanometer fiber. Our results provide mechanistic insights into how nucleosomes compact into higher-order chromatin fibers.
Journal information: Science
© 2014 Phys.org