Predicting the fate of stem cells
University of Toronto researchers have developed a method that can rapidly screen human stem cells and better control what they will turn into. The technology could have potential use in regenerative medicine and drug development. Findings are published in this week's issue of the journal Nature Methods.
"The work allows for a better understanding of how to turn stem cells into clinically useful cell types more efficiently," according to Emanuel Nazareth, a PhD student at the Institute of Biomaterials & Biomedical Engineering (IBBME) at the University of Toronto. The research comes out of the lab of Professor Peter Zandstra, Canada Research Chair in Bioengineering at U of T.
The researchers used human pluripotent stem cells (hPSC), cells which have the potential to differentiate and eventually become any type of cell in the body. But the key to getting stem cells to grow into specific types of cells, such as skin cells or heart tissue, is to grow them in the right environment in culture, and there have been challenges in getting those environments (which vary for different types of stem cells) just right, Nazareth said.
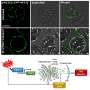
The researchers developed a high-throughput platform, which uses robotics and automation to test many compounds or drugs at once, with controllable environments to screen hPSCs in. With it, they can control the size of the stem cell colony, the density of cells, and other parameters in order to better study characteristics of the cells as they differentiate or turn into other cell types. Studies were done using stem cells in micro-environments optimized for screening and observing how they behaved when chemical changes were introduced.
It was found that two specific proteins within stem cells, Oct4 and Sox2, can be used to track the four major early cell fate types that stem cells can turn into, allowing four screens to be performed at once.
"One of the most frustrating challenges is that we have different research protocols for different cell types. But as it turns out, very often those protocols don't work across many different cell lines," Nazareth said.
The work also provides a way to study differences across cell lines that can be used to predict certain genetic information, such as abnormal chromosomes. What's more, these predictions can be done in a fraction of the time compared to other existing techniques, and for a substantially lower cost compared to other testing and screening methods.
"We anticipate this technology will underpin new strategies to identify cell fate control molecules, or even drugs, for a number of different stem cell types," Zandstra said.
As a drug screening technology "it's a dramatic improvement over its predecessors," said Nazareth. He notes that in some cases, the new technology can drop testing time from up to a month to a mere two days.
Professor Peter Zandstra was awarded the 2013 Till & McCulloch Award in recognition of this contribution to global stem cell research.
More information: High-throughput fingerprinting of human pluripotent stem cell fate responsiveness and lineage bias, DOI: 10.1038/nmeth.2684
Journal information: Nature Methods
Provided by University of Toronto