Biochemists identify protease substrates important for bacterial growth and development
(Phys.org) —Reporting this month in Molecular Microbiology, Peter Chien and colleagues at the University of Massachusetts Amherst describe using a combination of biochemistry and mass spectrometry to "trap" scores of new candidate substrates of the protease ClpXP to reveal how protein degradation is critical to cell cycle progression and bacterial development. The new understanding could lead to identifying new antibiotic targets.
As Chien (pronounced Chen) explains, to carry out fundamental life processes such as growing and dividing, cells must orchestrate, in time and location, the production and degradation of hundreds of protein substrates. Even in simple bacteria, protein degradation is critical for making sure these organisms can grow and respond to their environment properly.
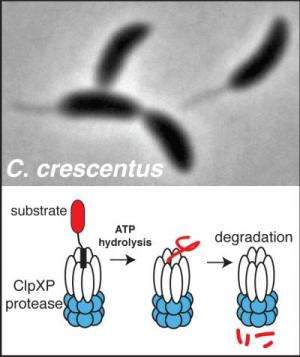
Scientists have known that a group of protein machines called energy-dependent proteases are responsible for the majority of this degradation, but what targets these machines recognize and how they do it has been unknown in many cases.
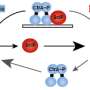
With the new series of experiments in the model bacteria Caulobacter crescentus in the Chien biochemistry and molecular biology laboratory, much more is now understood, he says. "We first generated a protease mutant that could recognize but not destroy its targets, acting as a 'trap' for protease substrates. After purifying this trap from living cells, we used mass spectrometry to identify proteins that were caught, finding over a hundred new candidate substrates. These targets covered all aspects of bacterial growth, including DNA replication, transcription and cytoskeletal changes."
Next, they focused on one of these new targets in detail, a protein called TacA. Caulobacter grow by making two different cell types every time they divide. TacA is responsible for making sure that one of these cell types forms properly.
"We used biochemistry and highly purified proteins to identify what parts of TacA were important for degradation by the ClpXP protease," Chien says. "We then made mutants of TacA that could not be degraded and found that when we expressed them in bacteria, these cells failed to properly develop into the correct cell types. Because developmental changes are essential for pathogenic bacteria to invade their host, these insights could potentially identify new antibiotic targets."
Journal information: Molecular Microbiology
Provided by University of Massachusetts Amherst