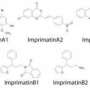
Clever use of high-resolution mass spectrometry allows rapid cataloging of sulfur-containing compounds in plant extracts

Plants are a rich resource of bioactive compounds, many of which have inspired therapeutic drugs. Yet countless plant compounds, potentially with medical uses, still remain to be identified. Kazuki Saito, Ryo Nakabayashi and colleagues from the RIKEN Plant Science Center have now developed a technique for rapidly cataloging subsets of compounds in plant extracts based on mass spectrometry data as a first step toward a fully automated system for cataloging novel plant compounds.
Mass spectrometry is an analytical technique that captures the mass spectrum of molecules in a sample, making it a powerful tool for identifying unknown compounds. Those compounds that contain certain 'heteroatoms' such as oxygen, nitrogen and sulfur produce a spectral 'fingerprint' that can be resolved in high-resolution mass spectra. Saito and his team hypothesized that this characteristic could be exploited to quickly catalog compounds in plant extracts.
To test their approach, the researchers profiled sulfur-containing compounds in onion extracts. Sulfur-containing compounds produce a pair of adjacent peaks in the mass spectrum—one 20 times stronger than the other—associated with the ratio of the two most common naturally occurring isotopes of the sulfur atom.
Using an ultrahigh-resolution, highly mass-accurate instrument called a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer, Saito and his colleagues were able to identify 67 sulfur-containing ions in the onion extract. Then, using isotope chemistry, they were able to establish the number of carbon atoms in each structure.
The researchers analyzed extracts from two sets of onions: one grown under a normal atmosphere, and another grown under an atmosphere in which the carbon in CO2 was replaced with the heavier carbon isotope, carbon-13. By measuring how much heavier a plant compound was when formed from carbon-13, the researchers could calculate its carbon count and so determine its complete atomic make-up. In a final step, the team established the full chemical structure of some of the sulfur-containing compounds by comparing their data with that of known compounds.
"We are now planning to develop an automated structural assignment system," says Nakabayashi. The researchers hope to automate this time-consuming process by incorporating an additional analysis technique called nuclear magnetic resonance (NMR), developing a database of high-quality mass spectrometry and NMR reference data, and establishing a computational algorithm for checking each compound against this database to establish its chemical structure. The researchers also plan to extend their existing technique to compounds containing other heteroatoms, such as oxygen, nitrogen, bromine and chlorine.
More information: Nakabayashi, R., et al. Combination of liquid chromatography−Fourier transform ion cyclotron resonance-mass spectrometry with 13C‑labeling for chemical assignment of sulfur-containing metabolites in onion bulbs. Analytical Chemistry 85, 1310–1315 (2013). dx.doi.org/10.1021/ac302733c
Journal information: Analytical Chemistry
Provided by RIKEN










.jpg)