Study uncovers how molecular parasite replicates, spreads ability to cause disease
(Phys.org)—An international team of researchers has uncovered how a molecular parasite responsible for playing a role in antibiotic-resistant disease, such as MRSA, can replicate and spread ability to cause disease, according to a new study published online this week in the Early Edition of the Proceedings of the National Academy of Sciences.
The findings may help researchers identify potential targets to block the spread of infection on the molecular level.
The study was conducted through a longstanding collaboration between researchers from the VCU School of Medicine, the New York University Medical Center and Instituto Valenciano de Investigaciones Agraias (CITA-IVIA) in Spain. The study builds on previous work by the team.
The VCU portion of the study was led by Gail Christie, Ph.D., professor of microbiology and immunology in the VCU School of Medicine, who has spent more than 30 years studying the interactions between bacterial viruses, or phages, and bacterial genomes to gain a better understanding of how disease can spread on the molecular level.
According to Christie, bacterial genomes are constantly changing due to the acquisition of new DNA that can bring in new traits. Genes that grant bacteria the ability to cause disease are often found on acquired pieces of DNA known as pathogenicity islands.
In the pathogen Staphylococcus aureus, which is a major cause of hospital- and community-acquired infections and increasingly resistant to antibiotics, some of the toxin genes that allow this organism to cause disease are carried on a novel, mobile pathogenicity island called a SaPI. SaPI is considered a molecular parasite.
SaPI "hijacks" a bacterial virus to spread among S. aureus bacteria, then directs it to preferentially package the SaPI DNA and carry it to new cells, said Christie. As the SaPI DNA is packaged and passed on, the SaPIs interfere with the growth of their helper phages.
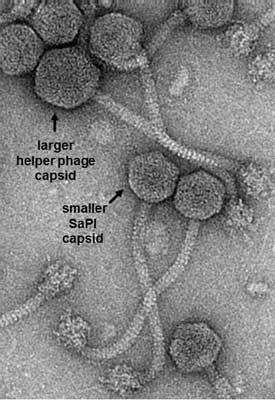
Previous work, by this team with additional collaborators, has focused on one aspect of this interference - the ability of the SaPIs to change the size of the capsid, or head, of the helper phage so that the smaller SaPI genome can be packaged but a complete helper phage genome cannot.
In this new study, the team found a second novel mechanism, which is the specific inhibition of packaging the helper phage DNA into the viral particles. Christie said the team also observed evidence for the existence of a third mechanism.
"Remarkably, different SaPIs use different combinations of these three mechanisms, depending on which particular helper phage they are exploiting," said Christie.
"Understanding these mechanisms will allow the identification of targets for blocking not only the spread of SaPIs, but of the helper phages themselves - many of which carry additional toxin genes on their own genomes," said Christie.
According to Christie, this is particularly important because clinical isolates of S. aureus carry both SaPIs and potential helper phages in their genomes, and the replication and spread of these elements can be induced by antibiotic treatment.
The general approach of investigating resistance to SaPI interference by identifying interference-resistant phage mutants was used first in Christie's lab. Those findings were reported in a research paper published in the journal Nature in 2010.
Journal information: Proceedings of the National Academy of Sciences , Nature
Provided by Virginia Commonwealth University