Loop the loop, DNA style: One- or two-way transcription depends on gene loops
(Phys.org)—In certain toy racecar tracks, sneaky players can flip a switch, trapping their opponents' vehicles in a loop of track. Cells employ a less subtle approach: they change the track's layout. In a study published online today in Science, scientists at the European Molecular Biology Laboratory (EMBL) and Oxford University discovered that, by forming or undoing gene loops, cells manipulate the path of the transcription machinery – which reads out instructions from DNA – controlling whether it moves along the genetic material in one direction or two.
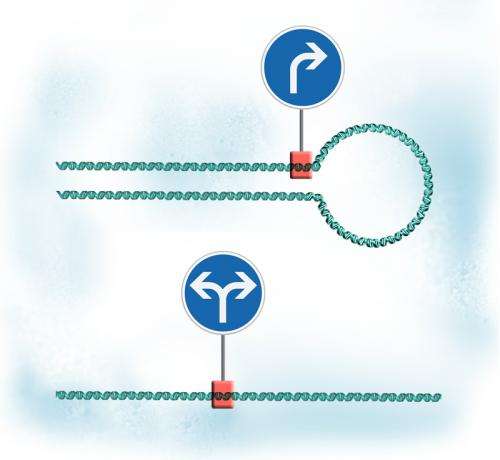
"We found that gene loops can turn bi-directional promoters into one-way systems," says Lars Steinmetz, who led the work at EMBL.
Three years ago, Steinmetz's lab discovered that when the transcription machinery lands on most genes' promoters – the sequences that mark where transcription should start – it moves not only along the gene, but also in the opposite direction, along the other 'lane' of DNA. But they were somewhat surprised that this didn't happen everywhere: some genes, it seemed, had promoters that worked only one-way. Nicholas Proudfoot's group at Oxford University, in the meantime, discovered that genes can bend into a loop, so that when the transcription machinery reaches the end of the gene, it finds itself back at the beginning and starts again, like a trapped toy car. Proudfoot's group also found that inactivating a specific protein prevented these gene loops from forming.
Looking throughout the whole genome of yeast cells, Steinmetz, Proudfoot and colleagues have now found that when genes with 'one-way' promoters can't form loops, transcription from those promoters becomes bi-directional. It seems that with no transcription 'cars' trapped in a loop, more are free to move in the opposite direction. And by doing so, they can affect other genes. So by forming or undoing gene loops, cells can control not only what happens to the looping genes themselves, but also adjust the spread of regulation throughout the genome.
The researchers would now like to understand how and when loops form, for instance in response to changes in a cell's environment, and how unlooping affects the transcription of short DNA sequences within the gene. And Steinmetz has another burning question: what happens in cases where a promoter activates transcription of two genes? "Are they each in a loop? What's going on there?" he wonders.
More information: Tan-Wong, S.M., Zaugg, J.B., Camblong, J., Xu, Z., Zhang, D.W., Mischo, H.E., Ansari, A.Z., Luscombe, N.M., Steinmetz, L.M., Proudfoot, N.J. Gene loops enhance transcriptional directionality. Published online in Science Express, 27 September 2012.
Abstract
Eukaryotic genomes are extensively transcribed, forming both messenger (m) and noncoding (nc) RNAs. ncRNAs made by RNA polymerase II (Pol II) often initiate from bidirectional promoters (nucleosome-depleted chromatin) that synthesise mRNA and ncRNA in opposite directions. We demonstrate that actively transcribed mRNA encoding genes by adopting a gene loop conformation, restrict divergent transcription of ncRNAs. Since gene loop formation depends on a protein factor (Ssu72) that co-associates with both promoter and terminator, its inactivation leads to increased synthesis of promoter-associated divergent ncRNAs, referred to as Ssu72 restricted transcripts (SRT). Similarly, inactivation of individual gene loops by gene mutation enhances SRT synthesis. We demonstrate that gene loop conformation enforces transcriptional directionality on otherwise bidirectional promoters.
Journal information: Science , Science Express
Provided by European Molecular Biology Laboratory