Numerical simulations reveal how protein function is affected by its naturally congested environment
The average cell of an organism is a packed environment, filled in particular with proteins—the workhorse molecules of the cell that are involved in almost every biochemical process taking place within it. A method to understand how this crowded environment affects the way that proteins function has been developed by Ryuhei Harada, at the RIKEN Advanced Institute for Computational Science in Kobe, and two RIKEN colleagues.
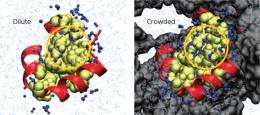
Although cells are well known to be crowded environments, studying protein function experimentally under crowded conditions is difficult, so researchers typically study proteins in dilute solutions instead. To get a more realistic picture of protein function as it actually takes place in nature, Harada and his colleagues use computer modeling.
“We carry out atom-level molecular dynamics simulations under crowded environments as a first step for understanding the cell,” Harada says. The researchers started by studying how the properties of the water molecules that surround the protein change when only a thin film of water separate one protein from its neighbor (Fig. 1). These properties can in turn profoundly influence the protein’s behavior.
“We were very surprised by the size of the effect that we calculated on water’s properties due to crowding,” Harada says. To measure the impact of these altered properties on protein function, the researchers modeled two small, simple proteins, each a sub-unit of a natural protein. They showed that crowding lowered the diffusion rates of the proteins through the cellular environment, which slowed protein dynamics.
Harada and colleagues also noted other effects. “Probably the most important finding from our study is the significant decrease in dielectric response of water in highly crowded environments,” Harada says. Water’s dielectric response relates to the way that it stores electrical charge. “The reduction of dielectric response directly impacts protein conformations and stabilities under crowded conditions.”
In particular, the reduced dielectric constant enhances the interactions between positively and negatively charged points in the protein structure that help to hold together its three-dimensional ‘secondary’ structure. The team found that the relationship is a linear one: the greater the crowding, the stronger the effect.
Harada and his colleagues are now investigating whether the observations that they made studying simple proteins will hold true for larger, more complex ones. They are also expanding their calculations to consider multiple proteins simultaneously. “We also want to develop general methodologies to simulate cellular environments based on this research,” Harada says.
More information: Harada, R., Sugita, Y. & Feig, M. Protein crowding affects hydration structure and dynamics. Journal of the American Chemical Society 134, 4842–4849 (2012)
Journal information: Journal of the American Chemical Society
Provided by RIKEN