Totally rad: Scientists create rewritable digital data storage in DNA
(Phys.org) -- Scientists from Stanford's Department of Bioengineering have devised a method for repeatedly encoding, storing and erasing digital data within the DNA of living cells.
Sometimes, remembering and forgetting are hard to do.
"It took us three years and 750 tries to make it work, but we finally did it," said Jerome Bonnet, PhD, of his latest research, a method for repeatedly encoding, storing and erasing digital data within the DNA of living cells.
Bonnet, a postdoctoral scholar at Stanford University, worked with graduate student Pakpoom Subsoontorn and assistant professor Drew Endy, PhD, to reapply natural enzymes adapted from bacteria to flip specific sequences of DNA back and forth at will. All three scientists work in the Department of Bioengineering, a joint effort of the School of Engineering and the School of Medicine.
In practical terms, they have devised the genetic equivalent of a binary digit — a "bit" in data parlance. "Essentially, if the DNA section points in one direction, it's a zero. If it points the other way, it's a one," Subsoontorn explained.
"Programmable data storage within the DNA of living cells would seem an incredibly powerful tool for studying cancer, aging, organismal development and even the natural environment," said Endy.
Researchers could count how many times a cell divides, for instance, and that might someday give scientists the ability to turn off cells before they turn cancerous.
In the computer world, their work would form the basis of what is known as non-volatile memory — data storage that can retain information without consuming power. In biotechnology, it is known by a slightly more technical term, recombinase-mediated DNA inversion, after the enzymatic processes used to cut, flip and recombine DNA within the cell.
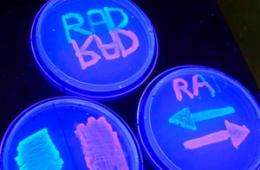
The team calls its device a "recombinase addressable data" module, or RAD for short. They used RAD to modify a particular section of DNA within microbes that determines how the one-celled organisms will fluoresce under ultraviolet light. The microbes glow red or green depending upon the orientation of the section of DNA. Using RAD, the engineers can flip the section back and forth at will.
They report their findings in a paper that will be published online May 21 in the Proceedings of the National Academy of Sciences. Bonnet is the first author of the paper, and Endy is the senior author.
To make their system work, the team had to control the precise dynamics of two opposing proteins, integrase and excisionase, within the microbes. "Previous work had shown how to flip the genetic sequence — albeit irreversibly — in one direction through the expression of a single enzyme," Bonnet said, "but we needed to reliably flip the sequence back and forth, over and over, in order to create a fully reusable binary data register, so we needed something different."
"The problem is that the proteins do their own thing. If both are active at the same time, or concentrated in the wrong amounts, you get a mess and the individual cells produce random results," Subsoontorn continued.
The researchers found it was fairly easy to flip a section of DNA in either direction. "But we discovered time and again that most of our designs failed when the two proteins were used together within the same cell," said Endy. "Ergo: Three years and 750 tries to get the balance of protein levels right."
Bonnet has now tested RAD modules in single microbes that have doubled more than 100 times and the switch has held. He has likewise switched the latch and watched a cell double 90 times, and set it back. The latch will even store information when the enzymes are not present. In short, RAD works. It is reliable and it is rewritable.
For Endy and the team, the future of computing then becomes not only how fast or how much can be computed, but when and where computations occur and how those computations might impact our understanding of and interaction with life.
"One of the coolest places for computing," Endy said, "is within biological systems."
His goal is to go from the single bit he has now to eight bits — or a "byte" — of programmable genetic data storage.
"I'm not even really concerned with the ways genetic data storage might be useful down the road, only in creating scalable and reliable biological bits as soon as possible. Then we'll put them in the hands of other scientists to show the world how they might be used," Endy said.
To get there, however, science will need many new tools for engineering biology, he added, but it will not be easy. "Such systems will likely be 10 to 50 times more complicated than current state-of-the-art genetic engineering projects," he said.
For what it is worth, Endy anticipates their second bit of rewritable DNA data will arrive faster than the first and the third faster still, but it will take time.
"We're probably looking at a decade from when we started to get to a full byte," he said. "But, by focusing today on tools that improve the engineering cycle at the heart of biotechnology, we'll help make all future engineering of biology easier, and that will lead us to much more interesting places."
More information: “Rewritable digital data storage in live cells via engineered control of recombination directionality,” Bonnet, J., Subsoontorn, P. & Endy, D. PNAS, dx.doi.org/10.1073/pnas.1202344109 (2012).
Journal information: Proceedings of the National Academy of Sciences
Provided by Stanford University Medical Center