Nanofluidics sorts DNA for cancer research
(Phys.org) -- Cornell nanotechnology researchers have devised a new tool to study epigenetic changes in DNA that can cause cancer and other diseases: a nanoscale fluidic device that sorts and collects DNA, one molecule at a time.
Epigenetics refers to chemical changes in DNA that do not alter the actual genetic code, but can influence the expression of genes and can be passed on when cells reproduce. One of the most important is DNA methylation, where methyl groups -- small structures of carbon and hydrogen -- are appended to DNA. Biologists study this by chemically precipitating out the methylated molecules, but these methods require large samples and often damage or throw away molecules they are supposed to find. Nanofluidics offers a way to select individual molecules out of tiny samples and collect them for further study.
The new device, developed in the lab of Harold Craighead, the Charles W. Lake Jr. Professor of Engineering, is described in the May 21 early online edition of the Proceedings of the National Academy of Sciences.
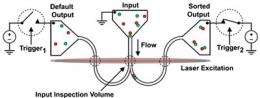
The process begins with a biochemical reaction that attaches a fluorescent tag to methylated DNA molecules. Then the sample is driven through a nanofluidic channel about 250 nanometers across -- so small that DNA molecules go through one at a time. Lasers illuminate the stream and cause fluorescence. When a fluorescing molecule goes by, a detector triggers a pulsed electric field that pushes the molecule to one side just before the channel splits into a Y. Methylated molecules go down one branch, everything else down the other.
"The color identity becomes a barcode for how the molecules are treated," explained Ben Cipriany, Ph.D. '12, lead researcher on the project, drawing an analogy to the methods used by the post office to sort packages on a conveyor belt. "Eventually we could use multiple colors, each representing a different epigenetic characteristic," he added.
The device reacts so swiftly that it can sort more than 500 molecules per minute, the researchers said. Fluorescent sorting is not new, they noted, but previously it has been done only with larger materials, such as nanoparticles or cells.
"We've made a miniaturized version that sorts individual molecules and works with a very tiny amount of input material," Cipriany said.
To test their method, the researchers observed fluorescence in each arm of the Y. They also collected the tiny sample of molecules that had been sorted out as methylated, amplified it using the PCR (polymerized chain reaction) method familiar to organic chemists and analyzed the resulting sample. False positives were limited to about 1-2 percent, which compares favorably with other sorting methods, they said.
The sorted output could be directed to a further microfluidic system for automated gene sequencing, the researchers suggested. The method also could be adapted to other molecule-separation tasks, they added.
The research was supported by the National Institutes of Health, Cornell Center for Invertebrate Genetics and the National Cancer Institute. Nanofabication was done at the Cornell NanoScale Science and Technology Facility, funded by the National Science Foundation.
Cipriany, who began this research as part of his Ph.D. studies at Cornell, is now at the IBM Semiconductor Research and Development Center in Hopewell Junction, N.Y.
Journal information: Proceedings of the National Academy of Sciences
Provided by Cornell University