Researchers demonstrate continuous and controlled translocation of DNA polymer through a nanopore
Research published this week in JACS (Journal of the American Chemical Society) shows continuous and controlled translocation of a single stranded DNA (ssDNA) polymer through a protein nanopore by a DNA polymerase enzyme. The paper by researchers at the University of California Santa Cruz (UCSC) provides the foundation for a molecular motor, an essential component of Strand Sequencing using nanopores. Researchers at UCSC are collaborating with the UK-based company Oxford Nanopore Technologies, developers of a nanopore DNA sequencing technology.
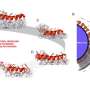
The new research advances previous work showing that DNA could be moved through a nanopore using a polymerase. DNA movement in the previous study was performed by a series of polymerases and required complex electronics for control. Improvements noted in the JACS paper include techniques to allow continuous ssDNA movement, giving an uninterrupted signal as the strand was moved through the nanopore in real time. The enzyme-nanopore construct was active and measurable in a constant electronic field without complex electronics.
Controlled initiation of the polymerase processing at the site of the nanopore-enzyme complex allowed sequential measurement of multiple ssDNA molecules using a single experimental setup . Furthermore the polymerase exhibited tenacious binding with the DNA polymer, unlike previous enzymes researched in similar conditions. These results demonstrate that qualities of the phi29 DNA polymerase are commensurate with a strand sequencing technology.
In the 'strand sequencing' method of nanopore DNA sequencing, ionic current through a protein nanopore is measured and current disruptions used to identify bases on a ssDNA polymer in sequence, as it translocates the pore. Two key challenges for this method are: engineering a nanopore to enable identification of individual bases when a ssDNA polymer spans the pore and a mechanism for controlling translocation of ssDNA at a consistent and appropriate speed to enable base identification through electronic measurements. Translocation techniques described in this paper are compatible with base identification technology being performed in the laboratories of Oxford Nanopore Technologies and its collaborators.
"This work with the phi29 polymerase has allowed us to make important progress on a key element of DNA strand sequencing," said investigator Professor Mark Akeson of the University of California, Santa Cruz. "While previous work showed that translocation control was possible in theory, this work shows that DNA translocation control is achievable in conditions that are compatible with an electronic sequencing technology. We look forward to further collaboration with Oxford Nanopore to realise this research."
"The 'strand sequencing' method of DNA sequencing using a nanopore has been studied for many years, but this paper shows for the first time that DNA can be translocated by an enzyme using methods that are consistent with a high throughput electronic technology," said Dr Gordon Sanghera, CEO of Oxford Nanopore. "We are excited by this work and its potential when coupled with additional recent developments in DNA base identification on DNA strands, the other critical element for strand sequencing."
More information: Processive Replication of Single DNA Molecules in a Nanopore Catalyzed by phi29 DNA Polymerase. Available online at pubs.acs.org/doi/abs/10.1021/ja1087612
Provided by Oxford Nanopore Technologies