This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Graph learning modules enhance drug-target interaction predictions
The identification of drug-target Interactions (DTIs) represents a pivotal link in the process of drug development and design. It plays a crucial role in narrowing the screening range of candidate drug molecules, thereby facilitating the reuse of drug, reducing the cost of drug development, and improving the efficiency of drug development.
Quantitative Biology has published an article titled "GCARDTI: Drug-target Interaction prediction based on the hybrid mechanism in drug SELFIES," which showed the utilization of drug SELFIES and the mechanism of mixing GCN and GAT demonstrated satisfactory performance.
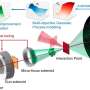
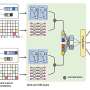
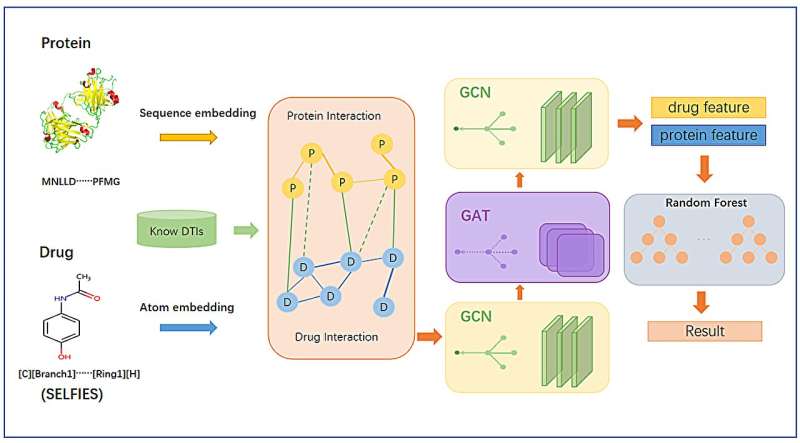
The GCARDTI model uses highly robust drug SELFIES combined with target sequences to build heterogeneous networks, and input them into a hybrid mechanism of graph learning module to capture high-dimensional features in the latent space of drugs and targets.
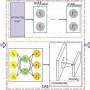
Graph convolutional neural networks automatically capture the structural information of drugs and target molecules by updating the feature vectors of adjacent atoms connected by chemical bonds. The attention module of the graph attention network is used to identify the contribution of the corresponding substructure to each drug and target.
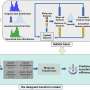
Finally, a layer graph convolutional neural network is used to aggregate the substructure information of each drug and target, and update its feature vector, so as to obtain low-dimensional and effective information of drugs and targets. By testing on data sets from two different sources, it is found that the GCARDTI model outperforms HIN2VEC, EVENT2VEC, HEER, GATNE, PGCNand, DTI-HETA in some aspects.
More information: Yinfei Feng et al, GCARDTI: Drug–target interaction prediction based on a hybrid mechanism in drug SELFIES, Quantitative Biology (2024). DOI: 10.1002/qub2.39
Provided by Higher Education Press