This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
A deep-learning framework for drug–drug interactions and drug–target interactions prediction
Exploring the biomedical interactions for chemical compounds and protein targets is crucial for drug discovery. Determining these drug–drug interactions (DDI) and drug–target interactions (DTI) not only reveals the potential synergistic effects of drug combinations and improves drug efficacy, but also contributes to drug reuse, reduces drug development costs, and improves drug development efficiency. Therefore, predicting interactions among drugs and drug targets is an important topic in the field of drug discovery.
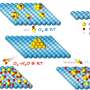
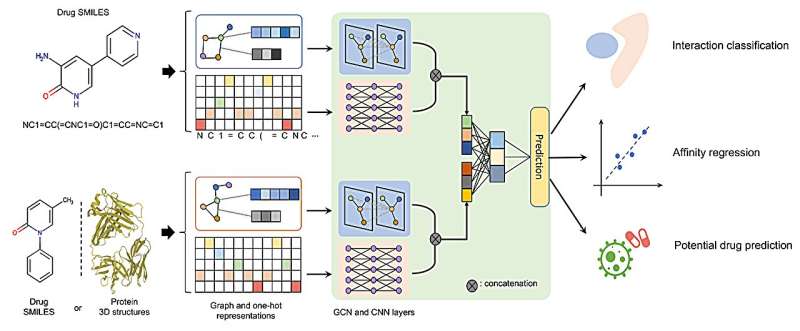
Quantitative Biology has recently published an article titled "DeepDrug: A general graph-based deep learning framework for drug–drug interactions and drug–target interactions prediction" that shows DeepDrug learns the comprehensive structure-based and sequence-based representations of drugs and proteins, achieving optimal performance across a range of tasks, by leveraging the residual graph convolutional networks and convolutional networks.
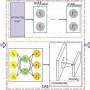
DeepDrug predicts drug/target interactions by combining sequence features and structural features, leveraging convolutional module and residual graph convolutional submodules, respectively. DeepDrug outperforms state-of-the-art methods in a series of systematic experiments, including binary-class DDIs, multi-class/multi-label DDIs, binary-class DTIs classification and DTIs regression tasks.
Furthermore, the structural features learned by DeepDrug display compatible and accordant patterns in chemical properties and drug categories, providing additional evidence to support the strong predictive capabilities of DeepDrug. As an application, DeepDrug is applied to discover the potential drug candidates against SARS-CoV-2, where 7 out of 10 top-ranked drugs are reported in the relevant literature.
More information: Qijin Yin et al, DeepDrug: A general graph‐based deep learning framework for drug‐drug interactions and drug‐target interactions prediction, Quantitative Biology (2023). DOI: 10.15302/J-QB-022-0320
Provided by Frontiers Journals