This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Genomewide prediction: A promising approach for improving russet formation in apple breeding programs
A research team has demonstrated that genomewide prediction is a promising approach for targeting russet formation in apple breeding programs. This method effectively predicts the russet trait, which is a complex quantitative trait controlled by many small-effect QTLs.
The study leveraged high-quality SNP data and observed moderate predictive abilities across various models. The study highlights the potential of genomewide prediction in targeting russet formation, thus facilitating the development of apple cultivars with improved fruit appearance and marketability.
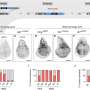
Appearance is a crucial attribute for fresh eating apples (Malus domestica Borkh.), with poor appearance causing significant financial loss as fruits are downgraded to the processed market. Russet formation, a superficial skin disorder, affects apple quality and storability, particularly in partially russeted cultivars. Phenotyping partial russet formation is challenging due to environmental influences and subjective assessments. Despite previous research on small-effect QTLs for russet, the genomewide prediction models utilizing fixed effects have not been studied in relation to russet formation.
A study published in Fruit Research on 12 June 2024, aims to evaluate genomewide prediction for partial russet formation in "Honeycrisp"-derived germplasm, hypothesizing improved predictive abilities with fixed-effect QTLs.
In this study, high-quality single nucleotide polymorphism data (947 SNPs) and three years of shoulder and lenticel russet formation data were leveraged. Moderate predictive abilities (mean r = 0.280.35) were observed across models and training sets. Specifically, the random effects model showed mean predictive abilities for shoulder russet formation as 0.33 for the All training set, 0.30 for the Honeycrisp training set, and 0.28 for the Minneiska training set.
The predictive abilities varied among families, with "Honeycrisp" × MN1964 showing the highest (r = 0.60) and "Honeycrisp" × AA44 the lowest (r = 0.12). For lenticel russet formation, predictive abilities were similar across training sets: 0.34 for the All training set, 0.30 for the Honeycrisp training set, and 0.35 for the Minneiska training set. The families with the highest predictive abilities were "Minneiska" × MN1702 (r = 0.53), the lowest predictive abilities were "Honeycrisp" × "WA 2" (r = 0.23).
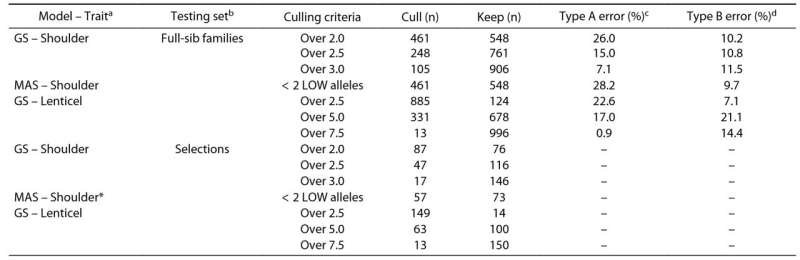
Additionally, the study found that the inclusion of previously detected QTLs as fixed effects in the models did not significantly enhance predictive abilities. Postdiction (retrospective) analysis revealed that genomewide prediction correctly categorized russet formation levels in 57% of advanced selections.
Sensitivity analysis indicated that marker-assisted and genomewide selection culled the same number of individuals for shoulder russet, but genomewide selection had fewer Type A misclassifications. For lenticel russet, genomewide selection retained 9%−92% of advanced selections, with no marker-assisted selection possible due to a lack of QTLs.
According to the study's lead researcher, Ashley A. Powell, "Genomewide prediction is a promising approach when targeting russet formation, a trait that cannot be phenotypically observed in offspring in apple breeding programs until they are past their juvenile phase."
In summary, this study utilized genomewide prediction to evaluate russet formation in apples, showing moderate predictive abilities (mean r = 0.28–0.35) across models and training sets. And fixed-effect QTLs did not significantly enhance predictions. Postdiction analysis confirmed the accuracy in categorizing russet levels. Looking forward, genomewide prediction offers promise for improving apple breeding efficiency, especially for complex traits like russet formation, by refining prediction models and incorporating comprehensive QTL data.
More information: Ashley A. Powell et al, Genomewide prediction to target russet formation in apple, Fruit Research (2024). DOI: 10.48130/frures-0024-0016
Provided by Maximum Academic Press