This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Low-cost depth imaging sensors achieve 97% accuracy in rapid plant disease detection
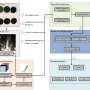
A research team has investigated low-cost depth imaging sensors with the objective of automating plant pathology tests. The team achieved 97% accuracy in distinguishing between resistant and susceptible plants based on cotyledon loss. This method operates 30 times faster than human annotation and is robust across various environments and plant densities.
The innovative imaging system, feature extraction method, and classification model provide a cost-efficient, high-throughput solution, with potential applications in decision-support tools and standalone technologies for real-time edge computation.
Selective plant breeding, which originated with the domestication of wild plants approximately 10,000 years ago, has evolved to address the challenges posed by climate change. Current breeding efforts focus on enhancing plant resilience to biotic and abiotic stresses, earlier germination, and improving nutritional and environmental values. However, the lengthy process of developing new varieties, which often takes up to 10 years, remains a significant hurdle.
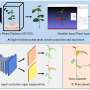
A study published in Plant Phenomics on 6 Jun 2024, investigates the effectiveness of Phenogrid, a phenotyping system designed for early-stage plant monitoring under biotic stress, addressing the issue of plant resistance to pathogens.
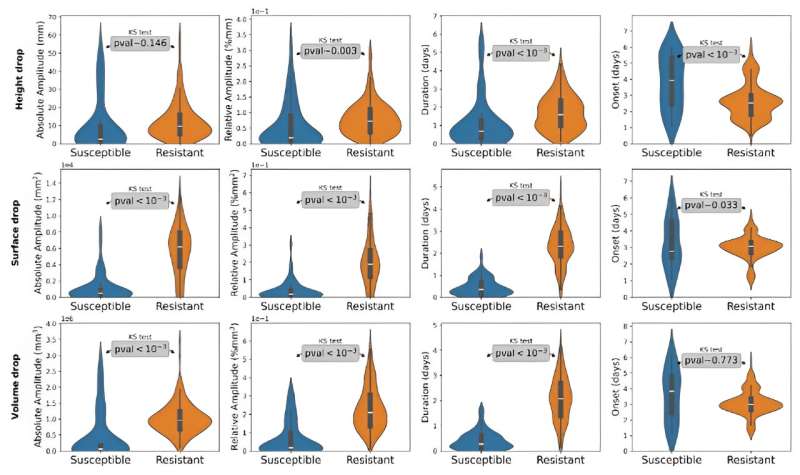
In this study, the extraction of spatio-temporal features, including absolute amplitude (Aabs), relative amplitude (Arel), and drop duration (D), proved to be an effective method for differentiating between susceptible and resistant plant batches.
The onset (O) feature demonstrated uniformity in susceptible plants, whereas resistant plants exhibited a consistent three-day onset, which correlated with cotyledon loss. Height signals were less effective, while surface and volume signals demonstrated pronounced contrasts between susceptible and resistant plants.
The results of the statistical tests demonstrated the significance of the majority of the extracted features in detecting cotyledon loss. This necessitated the use of a complex classifier in order to achieve efficient batch classification. The Random Forest model achieved the highest classification accuracy of 97%, accompanied by strong performance metrics (MCC: +91%).
The method demonstrated resilience to inoculation timing variability, maintaining performance with up to two hours of desynchronization. Furthermore, simulations indicated that reducing the number of plants per batch from 20 to 10 maintained classification performance while doubling throughput.
A visual analysis revealed that direct watering had an impact on classification accuracy, suggesting automated or subirrigation methods could further enhance performance. The method's efficacy extends to the segregation of other pathosystems, thereby demonstrating robust generalizability and potential for high-throughput plant pathology diagnostics.
The study's senior researcher, David Rousseau, asserts that the imaging system developed, when combined with the feature extraction method and classification model, provides a comprehensive pipeline with unparalleled throughput and cost efficiency when compared to the state-of-the-art.
The system can be deployed as a decision-support tool, but is also compatible with a standalone technology where computation is done at the edge in real time.
This study demonstrates the successful automation of plant pathology tests using low-cost depth imaging sensors, achieving an accuracy of 97% in distinguishing resistant from susceptible plants through cotyledon loss detection. The method is robust to variations in plant density and desynchronization, and thus significantly accelerates the processing time compared to that required for human annotation.
Future enhancements could include integrating additional imaging modalities and refining algorithms for broader applicability, promising a rapid, accurate, and cost-effective solution for improving crop resilience and productivity.
More information: Mathis Cordier et al, Affordable phenotyping system for automatic detection of hypersensitive reactions, Plant Phenomics (2024). DOI: 10.34133/plantphenomics.0204
Provided by TranSpread