This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Revolutionizing plant phenotyping: 3D plant shoot segmentation with deep learning
The quest for efficient 3D plant shoot segmentation, vital for extracting plant phenotypic traits, has driven the integration of deep learning with point clouds. Traditional 2D methods, while prevalent, encounter challenges in depth perception and structural determination. The emergence of 3D imaging has overcome these limitations, providing improved trait analysis.
However, the current research landscape is hampered by the need for extensive point-wise annotation in deep learning models, a process both costly and time-consuming. Addressing these issues, particularly through the development of extensive 3D datasets and exploration of weakly supervised learning models, stands as the critical frontier in advancing high-throughput plant phenotyping research.
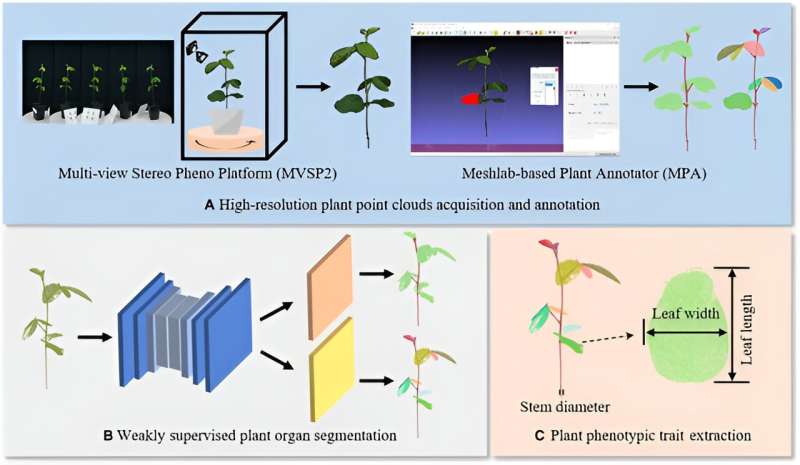
In a study titled "Eff-3DPSeg: 3D Organ-Level Plant Shoot Segmentation Using Annotation-Efficient Deep Learning " published in Plant Phenomics, researchers introduced the Eff-3DPSeg, a weakly supervised deep learning framework for plant organ segmentation. The methodology commences with the acquisition of point clouds from individual plants utilizing a Multi-view Stereo Pheno Platform (MVSP2). These point clouds are annotated using a Meshlab-based Plant Annotator (MPA), facilitating a weakly supervised learning process.
The framework was tested on a newly created, well-labeled large-scale soybean spatiotemporal dataset, capturing various growth stages. The framework's performance was compared with fully supervised methods on both soybean and tomato plants. The results demonstrated robust generalization and accuracy in stem-leaf segmentation, albeit with some misclassifications at junctions and leaf edges.
The method performed better on simpler plant structures and had a higher accuracy with more training data. Quantitative results indicated significant improvements over baseline methods in all metrics, with better performance in less supervised settings. For leaf instance segmentation, the framework achieved comparable results to full supervision but encountered challenges with dense leaves and limited samples. Improved segmentation accuracy was observed with increased supervision points.
The method excelled in extracting plant phenotypic traits, with some variations in performance between soybean and tomato plants based on the segmentation accuracy and quality of point clouds. Despite its promising results, the study faced limitations like data gaps and the need for separate training for different segmentation tasks. Future work aims to refine the framework, increase the variety of plant categories and growth stages, and improve the generality and diversity of the method.
In conclusion, the Eff-3DPSeg framework represents a significant step forward in 3D plant shoot segmentation. Its low-cost data acquisition, efficient annotation process, and robust segmentation capabilities hold great potential for enhancing high-throughput plant phenotyping and advancing smart agriculture.
More information: Liyi Luo et al, Eff-3DPSeg: 3D Organ-Level Plant Shoot Segmentation Using Annotation-Efficient Deep Learning, Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0080
Provided by Plant Phenomics