This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers reveal assembly of critical molecular machine that removes non-coding information from genes

One of the most striking features of human genes is that genetic information required to produce proteins is stored in a discontinuous form, wherein the coding information (exons) is punctuated with non-coding segments known as introns.
A new study from the Galej group at EMBL Grenoble contributes to a better understanding of the process by which this non-coding information is removed prior to protein synthesis.
In order to produce functional proteins, the cell must remove introns from precursors of messenger RNA molecules called pre-mRNAs—a process known as pre-mRNA splicing. In a way, splicing is similar to movie editing, where individual video clips are cut and stitched together to form a coherent story.
The splicing reaction is catalyzed by a large and dynamic molecular machine known as the spliceosome. The spliceosome surveys messenger RNA molecules to identify the beginning and the end of each coding segment and puts them together in a faithful way. This is of critical importance as any inaccuracies in this process may have drastic consequences for gene expression outcomes.
"Numerous genetic disorders are linked directly or indirectly to either mutations in spliceosome components or the sequences that it recognizes," said Wojtek Galej, Group Leader at EMBL Grenoble. "Therefore, investigation of the pre-mRNA splicing has a huge medical relevance and understanding of its fundamental mechanism may pave the way for novel therapies to improve human health."
The spliceosome is a very complex molecular machine, built from more than 100 proteins and five RNA molecules. These components are pre-assembled into five major building blocks known as small nuclear ribonucleoprotein particles (snRNPs, pronounced "snurps"). The Galej Group interrogated the structure of one of the building blocks of the spliceosome—the 20S U5 snRNP.
The largest building block of the spliceosome is called the tri-snRNP complex and is formed from the interaction of three different snRNPs—U4, U5, and U6. Although scientists know the structure of the mature tri-snRNP, it remains unclear how exactly its numerous components assemble.
Researchers from the Galej Group addressed this problem by analyzing the structure of 20S U5 snRNP, one of the intermediates in this assembly pathway. Although this intermediate was first isolated more than three decades ago, its structure has remained elusive until now.
The scientists purified their sample directly from human cells and visualized it using electron cryomicroscopy (cryoEM) and interpreted their data with the help of AlphaFold2, an artificial intelligence-based system for predicting protein structures. Their findings are published in the journal Nature Structural & Molecular Biology.
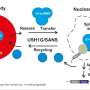
The researchers discovered that CD2BP2, one of the proteins in the tri-snRNP complex, acts as a molecular "chaperone," helping the protein components of the complex come together and assemble correctly. This protein is only present in the precursor form of the complex and leaves it as soon as it reaches its mature state.
To better understand the function of this factor, the team used CRISPR-Cas9 gene editing technology to create cell lines devoid of CD2BP2. With support from the Proteomics Core Facility at EMBL Heidelberg, the scientists found that in the absence of CD2BP2, snRNPs are produced less efficiently due to a potential roadblock in their assembly pathway.
"We started this project more than five years ago, and it has been a great collaborative effort of multiple people in the group looking at the problem from different angles. It is immensely satisfying to see final results coming together now," said Galej.
More information: Daria Riabov Bassat et al, Structural basis of human U5 snRNP late biogenesis and recycling, Nature Structural & Molecular Biology (2024). DOI: 10.1038/s41594-024-01243-4
Journal information: Nature Structural & Molecular Biology
Provided by European Molecular Biology Laboratory