This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers reveal structure and assembly mechanism of unique module in cellulosome
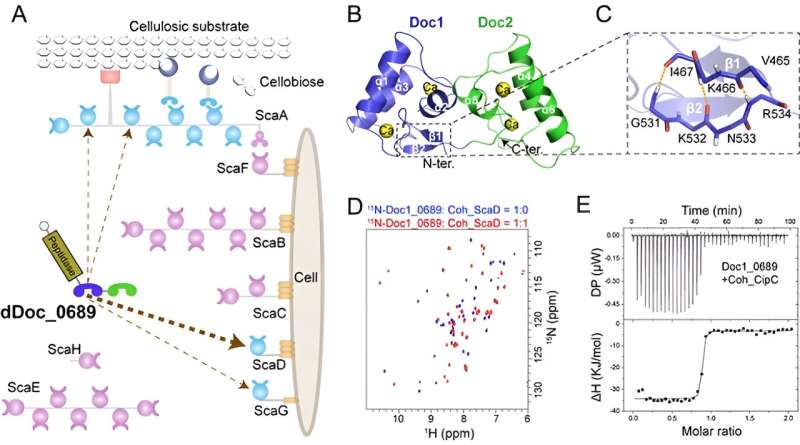
Cellulosomes are multi-enzyme complexes known for their efficient lignocellulose degradation, which is valuable in bioenergy technique development. The diverse composition and intricate assembly of cellulosomes give them exceptional substrate degradation capabilities, and deciphering their complex assembly mechanisms can provide a better understanding of their efficiency and promote their applications in bioenergy production.
Researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology of the Chinese Academy of Sciences have characterized the structure and assembly mode of a unique cellulosomal assembly module, the double-dockerin module, revealing the intricate complexity and diversity of cellulosome assembly and regulation. The results were published in Protein Science.
Cellulosomes are assembled by the interaction of scaffolding proteins (scaffoldins) and enzymes. The non-catalytic scaffoldins contain multiple tandem cohesin modules, whereas the cellulosomal enzymes are equipped with only a single dockerin module that allows specific binding to scaffoldins. Therefore, the structural complexity of the cellulosome is mainly determined by the number and type(s) of cohesins in the scaffoldins.
However, recent omics data reveal the presence of tandem double- and multiple dockerin modules in some cellulosome-producing bacteria. The role of these modules in cellulosomes needs to be elucidated.
In this study, the researchers have discovered a unique double dockerin module within a protease component in the cellulosome of Clostridium thermocellum.
Crystal and NMR structural analysis revealed that the double-dockerin module has two typical dockerin structures. However, a novel intramolecular clasp comprising anti-parallel β-strands was identified in the first dockerin module, and putative cohesin-binding residues in each module show certain variations compared to other conventional dockerin modules.
Further interaction studies demonstrated that only the first dockerin module is engaged in the assembly process, showing a preference for binding to the cohesin of a distinct cell wall-binding scaffoldin. This suggests that the double-dockerin module might play a role in regulating the assembly of cellulosomes.
In addition, the researchers have discovered that the double-dockerin module has a remarkable ability to bind to the heterologous cohesin module of Clostridium cellulolyticum. This finding represents a new example of the potential formation of "symbiotic cellulosomes" within a microbial community.
"This study shows that cellulosomes are much more complex than we thought, and unraveling this complexity is crucial for their application," said Prof. Feng Yingang, corresponding author of the study.
More information: Chao Chen et al, A cellulosomal double‐dockerin module from Clostridium thermocellum shows distinct structural and cohesin‐binding features, Protein Science (2024). DOI: 10.1002/pro.4937
Provided by Chinese Academy of Sciences




















