This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers reveal a unique, hitherto-unknown bacterial transcriptional promoter recognition mode
Researchers led by Prof. Zhu Ping from the Institute of Biophysics and Prof. Feng Yingang at the Qingdao Institute of Bioenergy and Bioprocess Technology, both under the Chinese Academy of Sciences, have revealed a unique, hitherto-unknown bacterial transcriptional promoter recognition mode by distinct σI factors in Clostridium thermocellum, both with respect to domain organization and binding mode to promoter DNA.
The study was published in Nature Communications on Oct. 13.
Lignocellulose can be efficiently degraded by a class of anaerobic clostridia represented by C.thermocellum through a secreted multienzyme complex, termed cellulosome, which is of great value in the development of bioenergy.
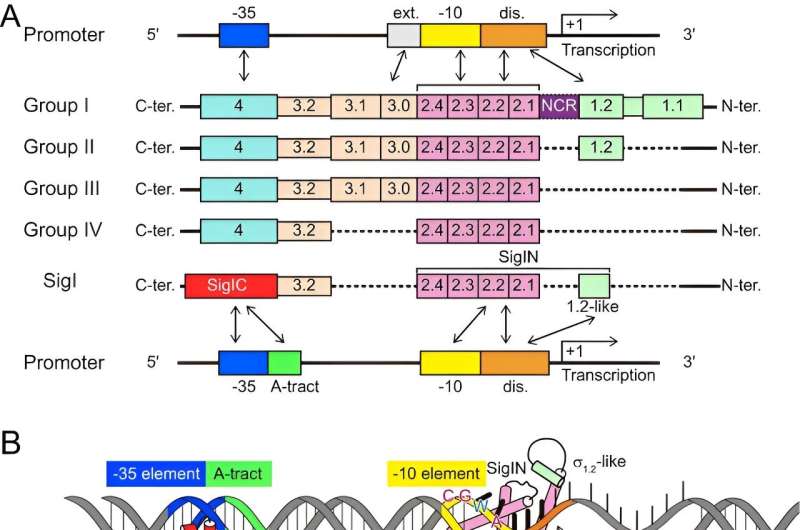
Bacterial σ factors are critical components of RNA polymerase (RNAP) holoenzymes for initiation of transcription by specifically recognizing DNA promoter regions. A single bacterium often contains multiple σ factors. However, the σI (SigI) in C. thermocellum exhibits lower homology to most σ factors and stands as a unique member within the bacterial σ factor family σ70.
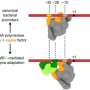
To reveal the molecular mechanism of how the SigI factors in C. thermocellum recognize promoters, the researchers prepared a ternary complex formed by SigI, RNAP, and promoter DNA, and determined the cryo-EM structures of RNAP-σ-promoter open complexes (RPo complexes) of two C. thermocellum σI complexes, i.e., RPo-SigI1 and RPo-SigI6 complexes, in the active state at 3.0Å and 3.3Å resolution, respectively.
Comparing the structures of these two RPo-SigI complexes with those of other transcriptional complexes, the researchers found that although the SigI-mediated transcriptional open complex presented an overall conserved architecture, SigI exhibited a distinctive promoter recognition mechanism among other σ factors in σ70 family.
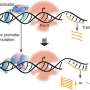
The C terminal domain of SigI (SigIC) presented a unique -35 element recognition mode. SigIC bound the DNA major groove of promoter -35 element through its helix-turn-helix (HTH) motif, which was rotated about 180° compared to the HTH binding in the major groove of σ4 domain of other σ factors in the σ70 family. In addition to the binding with the major groove, SigIC also inserted a conserved histidine into the DNA minor groove of -35 element, which is the conserved A-tract region of the promoter, representing a unique promoter recognition mode of SigI factors.
The recognition mode to the promoter -10 element by the N terminal domain of SigI (SigIN) also showed novel features: the SigIN domain had a conserved interaction mode with CGWA motif of -10 element while all transcription-bubble bases in -10 element interacting with SigIN were flipped out, which formed extensive protein-nucleic acid interactions with SigIN. These interactions may modulate the promoter activity and determine the differences in the transcription strength of SigI-dependent promoters of cellulosomal genes.
The study indicates that the transcriptional regulatory factor σI in C. thermocellum possesses a unique promoter recognition mechanism. This finding holds implications for understanding how C. thermocellum regulates the expression of cellulase genes and for the modification and application of its cellulases.
Additionally, the distinctiveness of σI factor in its promoter recognition mechanism enriches our understanding of the diversity of microbial transcriptional regulation.
More information: Jie Li et al, Structure of the transcription open complex of distinct σI factors, Nature Communications (2023). DOI: 10.1038/s41467-023-41796-4
Journal information: Nature Communications
Provided by Chinese Academy of Sciences