This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers discover new microRNA-regulated pathway to boost iPS cell-derived platelet production
A team of researchers led by Professor Koji Eto (Department of Clinical Application) recently discovered the importance of a microRNA-based regulatory mechanism for enhancing the quality and quantity of platelets generated by iPS cell-derived megakaryocytes. The findings are published in the journal Nature Communications.
Megakaryocytes (MKs) are traditionally recognized for their primary function of platelet production. However, new evidence in recent years suggests they may also possess immunomodulatory functions.
The research team recognized heterogeneity as a critical issue hindering the high-quality ex vivo manufacturing of iPS cell-derived platelets (iPSC-PLT) from immortalized MK progenitor cell lines (imMKCLs) and questioned whether they could identify specific imMKCL subpopulations based on differential microRNA (miRNA) activity.
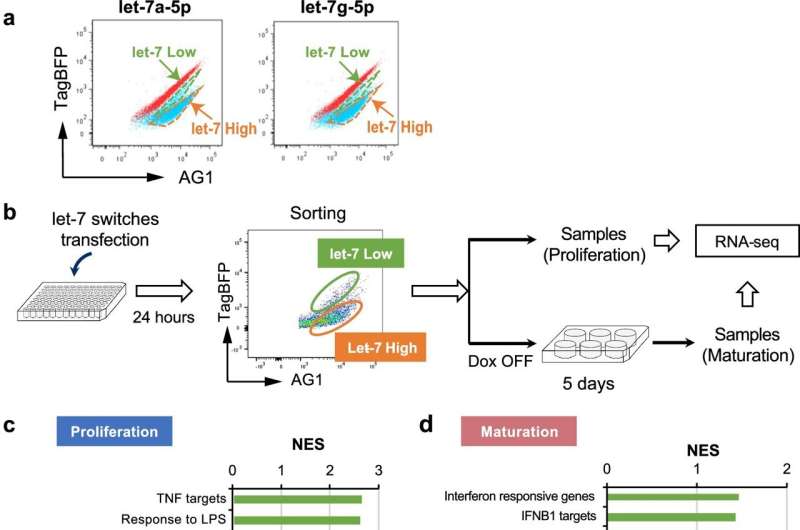
Employing miRNA switch technology developed by Professor Hirohide Saito (Department of Life Science Frontiers), the researchers screened a library of more than 250 miRNA switches and identified 24 active miRNAs in imMKCLs. From these active miRNAs, they chose to focus on let-7a-5p and let-7g-5p miRNAs because they observed distinct imMKCL subpopulations with either high or low activity of these miRNAs.
The researchers next separated imMKCLs into let-7 high and low subpopulations to examine their gene expression patterns during proliferation or maturation by bulk RNA sequencing (RNA-seq). Notably, they found immune-related genes were enriched in both let-7 low proliferating and mature imMKCL subpopulations, characteristic of MK progenitor cells with immunomodulatory functions.
The researchers obtained similar results when they examined the transcriptome of let-7 low and let-7 high CD34+ hematopoietic progenitor cells (HPCs) derived from human embryonic stem cells, thus revealing that such properties (thrombopoietic vs. immunomodulatory MK progenitor cells) are determined as early as the HPC stage.
When they further examined imMKCL heterogeneity at the single-cell (sc) level by scRNA-seq, five transcriptomically distinct cell subpopulations were identified, two primarily comprised of let-7 low imMKCLs. An in-depth examination of upregulated genes in these two cell clusters suggests they likely represent a subset with functional duality (thrombopoietic and immunomodulatory) and an immunomodulatory MK progenitor subset.
By treating imMKCLs with a miRNA let-7a-5p inhibitor, the research team observed significant upregulation of immune-related genes, such as PF4, PPBP, ISG15, and IFIT3, in the let-7 low imMKCL cell clusters. Furthermore, let-7 inhibition enhanced the release of IL-8, a proinflammatory cytokine, by imMKCLs in response to immune stimulation, thus demonstrating the crucial role of let-7 in determining the fate of immunomodulatory imMKCLs.
The researchers continued their investigation to understand how let-7 regulates MK fate determination by searching for candidate upstream regulators responsible for orchestrating the transcriptomic changes unique to the let-7 low imMKCL cell clusters via bioinformatic analysis.
Notably, while regulators associated with myeloid development, such as GATA1 and KLF2, were predicted to be active in thrombopoietic imMKCLs, IRF7 and IRF3, known virus-inducible gene activators, were predicted to be activated in immunomodulatory imMKCLs.
A total of 20 upstream regulators were predicted to be active in both cell clusters, of which CUX1 and RALB were upregulated upon let-7 inhibition. Remarkably, whereas CUX1 overexpression did not affect IRF7, ISG15, and IFIT3 expression, RALB overexpression significantly upregulated those interferon signaling genes. Yet, RALB overexpression interestingly did not affect thrombopoiesis-related genes, suggesting that it acts downstream of let-7 miRNA specifically to regulate immune-related genes in immunomodulatory imMKCLs.
Based on this new knowledge about let-7-mediated effects in imMKCLs, the research team demonstrated that both genetic and pharmacologic RALB inhibition could improve imMKCL proliferation and platelet production.
Through this study, the researchers uncovered a previously unknown let-7-RALB regulatory axis, which will no doubt contribute to the enhanced industrial-scale manufacturing of iPSC-PLTs for clinical applications in the future.
More information: Si Jing Chen et al, A let-7 microRNA-RALB axis links the immune properties of iPSC-derived megakaryocytes with platelet producibility, Nature Communications (2024). DOI: 10.1038/s41467-024-46605-0
Journal information: Nature Communications
Provided by Kyoto University