This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
How to track important changes in a dynamic network
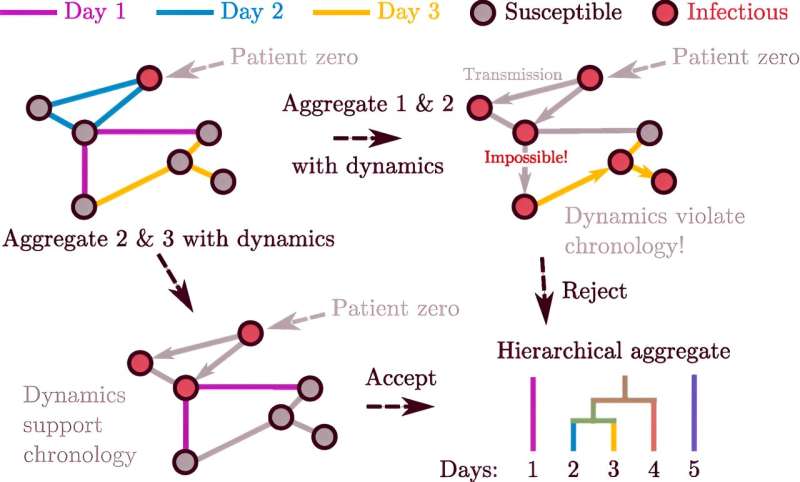
Networks can represent changing systems, like the spread of an epidemic or the growth of groups in a population of people. But the structure of these networks can change, too, as links appear or vanish over time. To better understand these changes, researchers often study a series of static "snapshots" that capture the structure of the network during a short duration.
Network theorists have sought ways to combine these snapshots. In a new paper in Physical Review Letters, a trio of SFI-affiliated researchers describe a novel way to aggregate static snapshots into smaller clusters of networks while still preserving the dynamic nature of the system. Their method, inspired by an idea from quantum mechanics, involves testing successive pairs of network snapshots to find those for which a combination would result in the smallest effect on the dynamics of the system—and then combining them.
Importantly, it can determine how to simplify the history of the network's structure as much as possible while maintaining accuracy. The math behind the method is fairly simple, says lead author Andrea Allen, now a data scientist at Children's Hospital of Philadelphia.
"We're really excited about being able to share it, and it's a wonder that nobody else has published this exact idea in the last decade," says Allen. She collaborated with SFI Professor Cris Moore, a physicist and mathematician, and Laurent Hébert-Dufresne, a complexity scientist at the University of Vermont and a former SFI James S. McDonnell Foundation Fellow.
In the published paper, the method doesn't appear complicated; in reality, it evolved over years both at and beyond SFI. The collaboration began in 2015 when Allen, then an undergraduate math major, visited SFI for a month in the winter and then, in the summer of 2016, returned to participate in the Research Experiences for Undergraduates program (now called the Undergraduate Complexity Research program).
Hébert-Dufresne had obtained a large dataset, acquired from satellite phone data, that used cell phone "pings" to show how people moved around. He was interested in finding communities, but he also wanted to be able to measure whether different communities required different data resolution.
"For instance, should epidemic surveillance systems be uniform across communities when we know that different communities have different behaviors?"
That question led to even more: "At what level can we aggregate this while still maintaining the differences? And how do we know?" Allen asks. "We don't want to lose the integrity of the network we're trying to study."
They brought in Moore to brainstorm ideas for how to know which differences were important to the overall structure, and which ones were less important. Then they shelved the project after a while.
Allen left academia to become a software developer and Hébert-Dufresne started his own research group in Vermont. But it would be a short hiatus. Two years later, Allen joined Hébert-Dufresne's group in Vermont as a graduate student and they picked up where they'd left off.
"We always said, 'let's wrap this up now,'" Allen says. "This kind of became a joke for eight years."
In the final push, the researchers identified a straightforward way to approximate error—and to use it in successive combinations of pairs of networks. In the paper, the researchers use the spread of disease as a measuring stick to assess and validate the method.
"Suppose there's a pandemic," says Moore. If two people—Alice and Bob—get together, and then two other people—say Bob and Charlene—get together, then the disease could spread from Alice to Charlene but not the other way around. The order of these links matters, which means it's misleading to combine them into one snapshot (and treat them as though they're simultaneous).
The new method borrows an idea from quantum mechanics to identify these kinds of errors. In that field, the "commutator" can reveal how much order matters in computations involving things like energy and momentum. In the new application, the researchers used a commutator to decide how much order matters, and when it's accurate to combine snapshots.
"This lets us simplify the history of the network's structure as much as possible while maintaining accuracy," says Moore. It also points to a way to tame an enormous, unwieldy dataset into a smaller, manageable set of networks.
Allen says it could be extended to other dynamic systems like the spread of information over a social media network.
More information: Andrea J. Allen et al, Compressing the Chronology of a Temporal Network with Graph Commutators, Physical Review Letters (2024). DOI: 10.1103/PhysRevLett.132.077402
Provided by Santa Fe Institute