This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
AI helps provide the most complete map of interactions key to bacterial survival
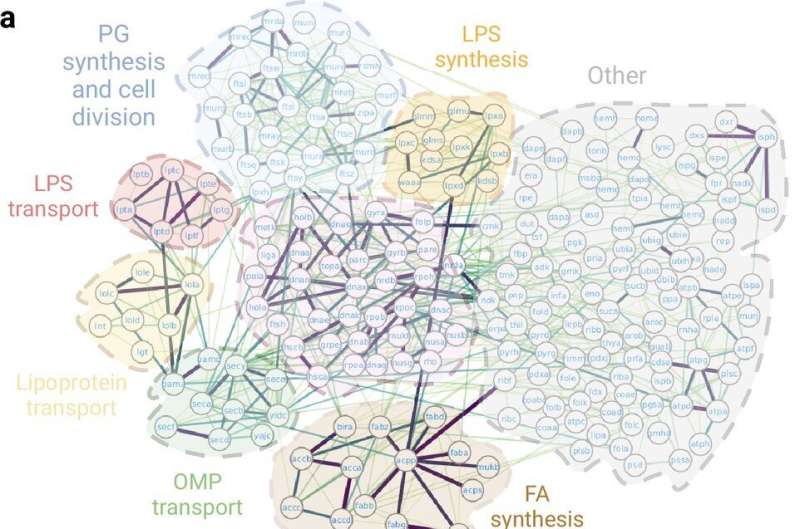
UAB researchers have produced the most complete map of the bacterial essential interactome, that is, how proteins combine and interact to perform functions essential for their survival. The research, published in the journal eLife, used the artificial intelligence tool AlphaFold to predict and model more than 1,400 interactions. The results have revealed previously unknown details of these mechanisms and offer potential targets for the development of new antibiotics.
Bacteria carry out many functions that are key to their survival, such as the production of the energy they need, DNA replication and cell division to reproduce, or the synthesis of their cell membrane to protect themselves and interact with the environment, among others. All these processes involve complexes that require the coordinated action of a set of proteins that are essential: without them, the processes do not take place and the bacteria die.
Therefore, knowing in detail how these basic processes are regulated, which proteins are involved and how they interact is essential to understanding the mechanisms of bacterial growth, reproduction and survival.
Experimental techniques undertaken so far have allowed the identification of millions of interactions between proteins and thousands of structures of these proteins, but these are raw data that give a large number of false positives; interactions that, in reality, have no value.
With recently developed artificial intelligence models such as AlphaFold2, it has been possible to obtain protein structures with an accuracy similar to experimental methods, and to differentiate between genuine protein–protein interactions and false interactions (false positives).
Researchers from the Department of Biochemistry and Molecular Biology at the Universitat Autònoma de Barcelona have used the AlphaFold2 artificial intelligence model to predict the set of protein–protein interactions that are essential for the survival of bacteria, a total of 1,402 possible interactions that make up the most complete map of what is called the bacterial essential interactome.
All these interactions expand our knowledge of the mechanisms of action that bacteria need to survive and allow us to identify which protein–protein interactions may be targets for the development of new antibiotics.
"We have obtained a map of the bacterial essential interactome in which all the interactions that are essential for bacteria to live and multiply are collected. We have structurally characterized these interactions using new artificial intelligence tools, specifically AlphaFold," explains UAB lecturer Marc Torrent, director of the research. "We believe that these structures are a reference for the development of new antibiotics, since molecules that can inhibit these interactions would behave like antibiotics with unusual mechanisms of action."
Bacterial activity involves between 4,000 and 5,000 proteins. This set is called the bacterial proteome, giving rise to an interactome that could have as many as 20 million possible interactions. But it is estimated that the interactions that take place in a species, for example, in the bacterium Escherichia coli, are limited to about 12,000. And not all of these interactions are essential for the survival of the bacterium.
To distinguish essential interactions, researchers considered only those where the two proteins that interact to form the complex are present in at least two different bacterial species. With these filters and the help of the artificial intelligence model AlphaFold2, researchers obtained a set of 1,402 essential protein-protein interactions.
Excellent predictive power of artificial intelligence
To test the reliability of AlphaFold2, the research team compared its predictions with 140 protein–protein interactions that had been obtained experimentally beforehand. The result was a predictive power that the authors describe as excellent, as 113 of these experimental interactions (81%) were predicted by the AI very accurately.
The researchers believe that many of the protein–protein interaction complexes that can be found in experimental databases could be false positives.
New, previously unknown essential protein complexes
The researchers highlight the discovery, using this method, of a set of previously unknown protein–protein interactions that act in nine essential processes: fatty acid biosynthesis in the cell membrane, lipopolysaccharide synthesis in the outer membrane, lipid transport, protein and lipoprotein transport in the outer membrane, cell division, maintenance of the elongated shape in bacilli, DNA replication for bacterial reproduction, and ubiquinone synthesis.
A detailed understanding of the structure of these newly discovered protein complexes provides new insights into the molecular mechanisms involved in these vital bacterial processes and paves the way for the development of new antibiotics.
More information: Jordi Gómez Borrego et al, Structural assembly of the bacterial essential interactome, eLife (2024). DOI: 10.7554/eLife.94919
Provided by Autonomous University of Barcelona





















