This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers use scattering function to analyze movement patterns of E. coli
In a joint effort with various international institutions, researchers from the University of Innsbruck have described the movement patterns of the bacterium Escherichia coli. To do so, they used an engineered bacterial strain, experiments under the microscope and complicated functions.
Escherichia coli is one of the world's best-known bacteria. Not only is it found in abundance in our intestines, it is also a favorite model organism in research. Under the microscope, E. coli bacteria can be recognized as a bunch of rods that are always on the move.
The exact form of this movement has now been investigated by several teams in an international research project. Scientists cultivated a new strain of bacteria whose movements can be controlled and compared experimental data with a physical model that describes the movement patterns of E. coli over long periods of time. This model was confirmed by the experimental study.
The comparison of experiments and theory using an intermediate scattering function is the achievement of Tyrolean scientist Christina Kurzthaler. This work began as part of her doctoral thesis in Thomas Franosch's research group at the Institute of Theoretical Physics at the University of Innsbruck.
Kurzthaler is one of the first authors of the study and now heads her own research group at the Max Planck Institute for the Physics of Complex Systems in Dresden. Her work has just been published as an Editor's Pick in Physical Review Letters.
Swimming and tumbling
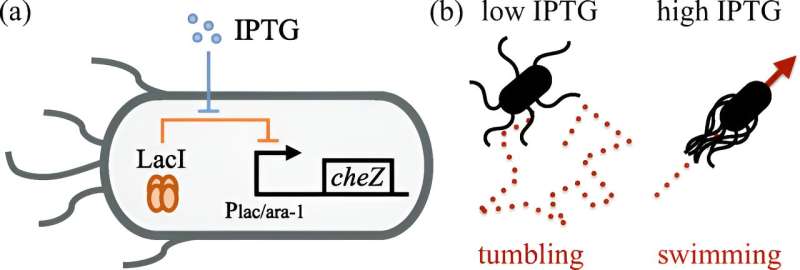
In biophysics, the movement of E. coli is described by the "run-and-tumble" model. With the help of many small flagella, the bacteria swim in a certain direction. At a certain point, the movement turns into tumbling and the bacteria change direction. This characteristic behavior has been known for a long time, but could not be described precisely until now, as it was hardly possible to measure how long the bacteria swim before they begin to tumble.
"E. coli bacteria swim in a solution, which means that they move very quickly in a three-dimensional space. This movement is difficult to measure because it requires a lot of data," says Kurzthaler. "Tracking individual bacteria over a long period of time is very time-consuming and requires special experimental instruments."
The tumbling behavior is very important for the bacterium. E. coli uses it to search for food or to escape from toxic substances. "Knowing this behavior in detail opens up many new experimental possibilities," says Kurzthaler.
Confirmation through scattering function
In order to accurately characterize the movement processes, partner groups at Chinese research institutions developed an engineered strain of E. coli in which the frequency of tumbling can be reduced or increased when grown in certain chemical solutions.
A group at the University of Edinburgh conducted experiments on these bacteria and took microscopic images of the entire bacterial population at multiple time points. The researchers in Innsbruck then analyzed the collected data by using an intermediate scattering function developed by Kurzthaler, which indirectly measures the distribution of the bacteria in space and time and provides information about their dynamics.
This made it possible to calculate a variety of data over long periods of time, for example, the speed of the bacteria and the duration of their tumbling. The result was a detailed description of the movement of E. coli bacteria in three-dimensional space.
"The run-and-tumble model itself is not new," says Franosch. "Our approach of calculating a movement in space with numerical solutions on the computer is. This is already common practice in other fields such as solid-state physics, but it is an innovation in biophysics.
"And the result is something fascinating: that this really simple run-and-tumble model perfectly describes the movement of bacteria. We checked it with our complicated function and couldn't find any deviation. In the field of biophysics, it is quite amazing that a theoretical model can be confirmed so precisely by experiments."
More information: Christina Kurzthaler et al, Characterization and Control of the Run-and-Tumble Dynamics of Escherichia Coli, Physical Review Letters (2024). DOI: 10.1103/PhysRevLett.132.038302
Journal information: Physical Review Letters
Provided by University of Innsbruck