This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Breakthrough in designing complicated all-α protein structures
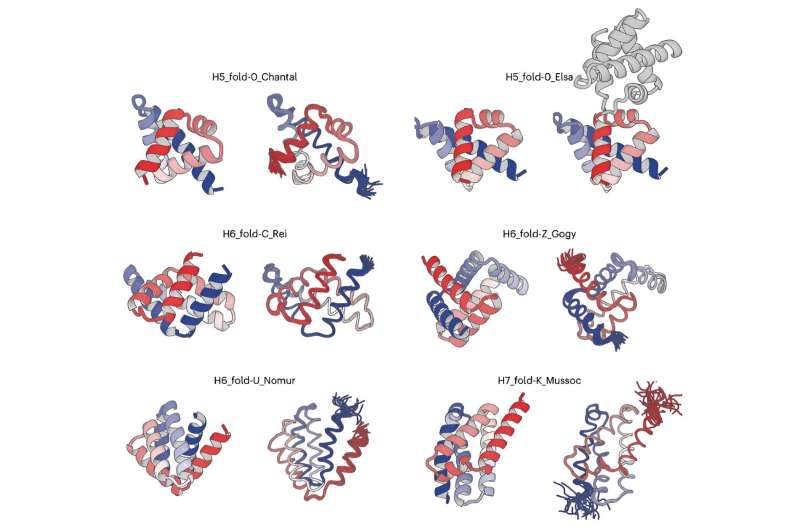
A team of researchers has developed an innovative method to design complicated all-α proteins, characterized by their non-uniformly arranged α-helices as seen in hemoglobin. Employing their novel approach, the team successfully created five unique all-α protein structures, each distinguished by their complicated arrangements of α-helices. This capability holds immense potential in designing functional proteins.
This research has been published in the journal Nature Structural & Molecular Biology.
Proteins fold into unique three-dimensional structures based on their amino acid sequences, which then dictate their function. Although significant progress has been made in de novo protein design, the ability to design complicated all-α proteins, where α-helices are non-parallelly arranged within the three-dimensional structures, was lacking.
"Artificially designed proteins mostly show simple structures, but nature presents us with complicated 'designs,'" said Prof. Nobuyasu Koga of Exploratory Research Center on Life and Living Systems (ExCELLS) at National Institutes of Natural Sciences (NINS). This gap drove the team to seek a method to design such complicated all-α proteins.
The team began by examining structures deposited in the Protein Data Bank (PDB) and identified 18 typical helix-loop-helix motifs. They then demonstrated that a broad spectrum of all-α protein tertiary structures, ranging from simple to complicated, can be computationally generated by combining these identified typical motifs and canonical α-helices.
"It is surprising that such a diverse set of all-α protein structures can be generated simply by combining typical or canonical components of naturally occurring proteins," said Dr. Koya Sakuma, a former Ph.D. student in SOKENDAI (The Graduate University for Advanced Studies).
The team selected five unique all-α proteins, each with five or six α-helices and distinguished by their complicated spatial arrangements, from the computationally generated structures. They then carried out de novo design of amino acid sequences that are capable of folding into the selected five all-α protein structures.
The experimental results were remarkable. "The structures that emerged from structure determination process exhibit the complicated shapes, which perfectly matched computationally designed structures," said Dr. Naohiro Kobayashi, a senior research fellow at RIKEN.
With this developed method, it is now possible to create all-α proteins with complicated shapes. Functions of proteins inherently depend on their three-dimensional structures.
If we can design proteins with complicated shapes, it increases the potential to construct functional sites within them. This breakthrough research will pave the way for the creation of new functional proteins, which will contribute to health care and life sciences.
More information: Design of complicated all-α protein structures, Nature Structural & Molecular Biology (2024). DOI: 10.1038/s41594-023-01147-9 www.nature.com/articles/s41594-023-01147-9
Journal information: Nature Structural & Molecular Biology
Provided by National Institutes of Natural Sciences