This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Expansin genes shown to drive heteroblastic leaves in Ceratopteris chingii
Heteroblasty is a developmental trajectory event in which plants undergo rapid ontogenetic changes in multiple traits, as exemplified by the transition from distinct juvenile to adult leaves. Heteroblastic leaves allow plants to adapt to environmental heterogeneity and serve as a prime example of adaptive evolution.
Previous studies have uncovered some of the underlying differences in heteroblasty in aquatic seed plants, but the mechanism behind sterile-fertile leaf dimorphy remains relatively unexplored in aquatic ferns.
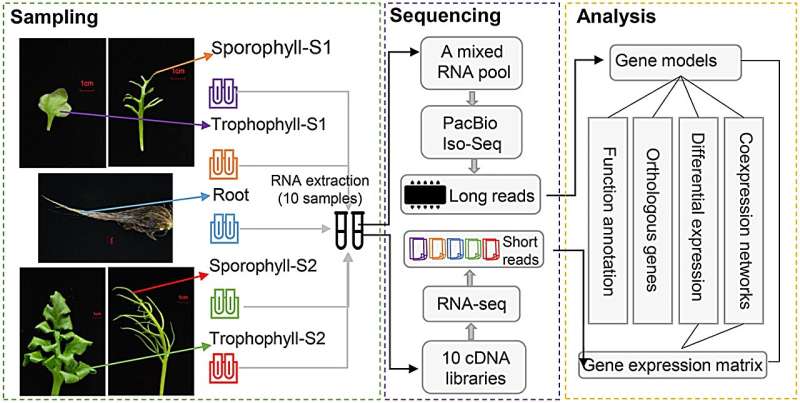
To understand the molecular mechanism regulating the heteroblastic leaves in Ceratopteris chingii, researchers from the Wuhan Botanical Garden of the Chinese Academy of Sciences (CAS), Hubei Ecology Polytechnic College, and Ghent University examined the transcriptome datasets between sporophylls and trophophylls using PacBio full-length sequencing and Illumina RNA-seq.
Comparative analysis revealed differentially expressed genes involved in reproduction and cell wall composition pathways. Of these, expansion genes account for the majority of significantly enriched Gene Ontology terms.
The results were published in BMC Biology, titled "Expression divergence of expansin genes drive the heteroblasty in Ceratopteris chingii."
Reconstruction of the expansion gene phylogeny revealed four distinct phylogenetic groups of expansion genes were distinguished in 19 plant species, ranging from green algae to seed plants. Co-expression analysis showed that the expression behaviors were highly divergent both within and between species.
In short, specific regulatory interactions and associated expression patterns of expansion genes played an important role in heteroblastic leaves of Ceratopteris chingii.
More information: Yue Zhang et al, Expression divergence of expansin genes drive the heteroblasty in Ceratopteris chingii, BMC Biology (2023). DOI: 10.1186/s12915-023-01743-7
Journal information: BMC Biology
Provided by Chinese Academy of Sciences