This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Forest polyploid breeding group analyzes the genetic regulatory network of poplar leaf development
As an important source organ, leaves are the main site of photosynthesis in plants, and analyzing the developmental process of leaves can help to improve the photosynthetic assimilation capacity of plants.
Plants of the genus Populus (Populus spp.) are widely used globally in the fields of environment, agroforestry and industry. Poplar is not only a horticultural greening species in modern cities, but also a model species for physiological and ecological studies.
Currently, studies on the developmental stages of poplar leaves have not been reported. Using high-throughput sequencing technology, the regular changes in gene and miRNA expression during the development of poplar leaves, constructing a dynamic transcriptome regulatory network of poplar leaf life cycle, and analyzing the core regulatory factors affecting leaf growth and development will help to understand the gene expression law of leaf development and provide effective reference for subsequent molecular design and breeding.
The article "Spatiotemporal miRNA and transcriptomic network dynamically regulate the developmental and senescence processes of poplar leaves," has been published in Horticulture Research.
The researchers found that the developmental process of poplar leaves is divided into five periods.
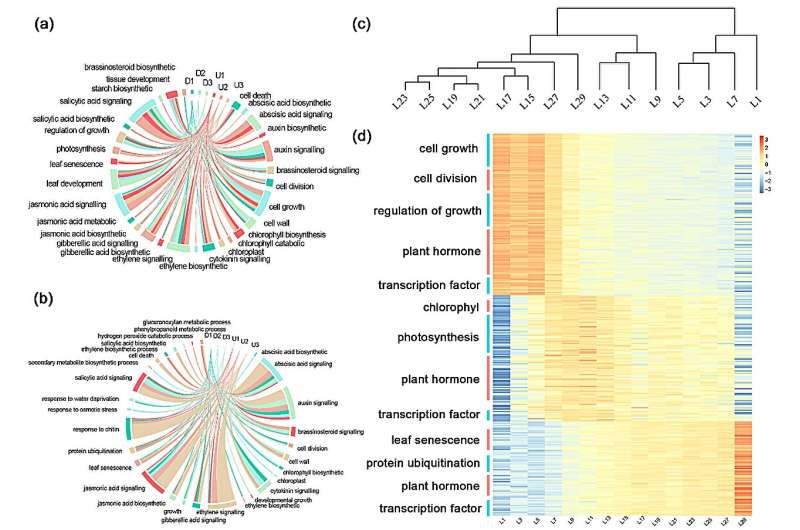
During the period of cellular genesis and functional differentiation of juvenile leaves (leaf position 1), the expression of genes related to cellular growth and division of leaves is higher; during the period of development of juvenile leaves and the initial formation of photosynthetic capacity (leaf positions 3-7), the expression of genes related to cellular growth and division decreases; during the period of the strongest photosynthetic capacity of functional leaves (leaf positions 9-13), photosynthesis and chlorophyll are reduced.
During the period of the strongest photosynthetic capacity of functional leaves (leaf positions 9-13), photosynthesis and chlorophyll are reduced), the expression of photosynthesis and chlorophyll metabolism pathway genes gradually increased; during the period of declining photosynthetic capacity of functional leaves (leaf position 15-27), the expression of photosynthesis-related genes decreased, the expression of senescence factors gradually increased, and the photosynthetic rate gradually declined.
In the period of senescent leaves (leaf position 29) the expression of senescence factors and protein ubiquitylization-related pathway genes was the highest, and the photosynthetic rate decreased to the lowest level at this time.
The expression of senescence factor and protein ubiquitination-related pathway genes was relatively highest during the senescence leaf period (29th leaf position), and the photosynthetic rate also dropped to the lowest level at this time.
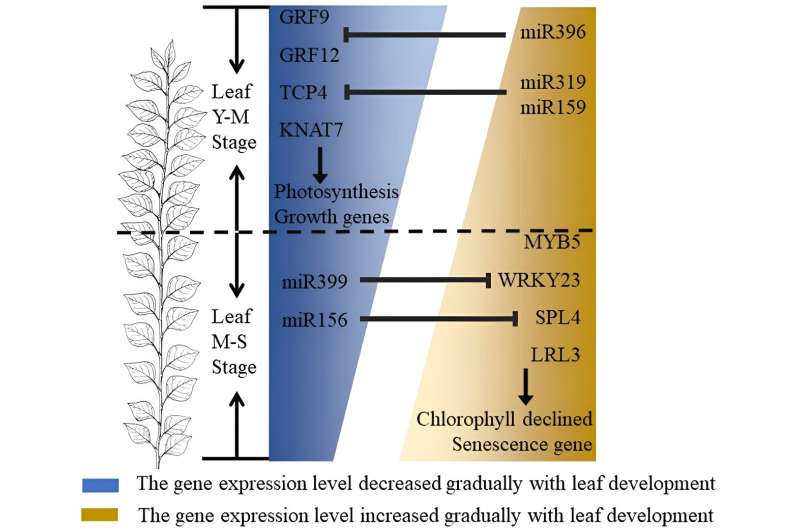
Therefore, miRNAs and transcription factors involved in the regulation of leaf growth and development at different periods showed period-specific expression patterns. Among them, GRF9, GRF12, KNAT7 and TCP4 have the role of promoting leaf growth and morphogenesis, and the expression of related transcription factors and their regulated functional genes gradually decreased as leaf development was regulated by miR396, miR159 and other factors.
While MYB5, SPL4, LRL3 and WRKY23 are involved in the regulation of leaf cell senescence, chlorophyll degradation and other biological processes, as leaf development is regulated by miR399, miR156 and other factors, the expression of related transcription factors and their regulated functional genes gradually increased with MYB5, SPL as the core of the regulatory network.
The key position of MYB5 in regulating pigment synthesis was established by constructing the gene expression regulatory network, and was verified by the phenotype of MYB5 overexpression lines.
The results help to deepen the understanding of the growth and development process of poplar leaves and its regulatory mechanism, and provide a new direction for molecular design breeding to slow down leaf senescence, increase the maximum photosynthetic rate of leaves, and grow the highest photosynthetic period.
More information: Kang Du et al, Spatiotemporal miRNA and transcriptomic network dynamically regulate the developmental and senescence processes of poplar leaves, Horticulture Research (2023). DOI: 10.1093/hr/uhad186
Provided by TranSpread