This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists develop open archive of plant images and related phenotypic traits
Plant images contain a wealth of information that reflects key phenotypic characteristics such as color, shape, growth, and health status of plants. High-throughput plant phenotypic collection technology has been widely applied in plant phenomics, generating numerous images and image-based traits (i-traits) data. These data serve as important resources for various agricultural applications, including germplasm screening, plant pests and disease identification, and agronomic traits mining.
Building a plant image and related trait data management platform provides centralized management, analysis, and sharing of plant images and related trait data. The platform not only facilitates data querying, access, interoperability, and reuse but also contributes to the standardization of image meta-information and phenotypic data. Such endeavors provide a crucial support platform for the application of plant phenomics driven by smart agriculture.
The Beijing Institute of Genomics of the Chinese Academy of Sciences (China National Center for Bioinformation) and the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences have jointly developed the Open Archive of Plant Image and Traits (OPIA), which provides a public service for domestic and foreign researchers to submit and share plant image and trait data. The study was published online in Nucleic Acid Research.
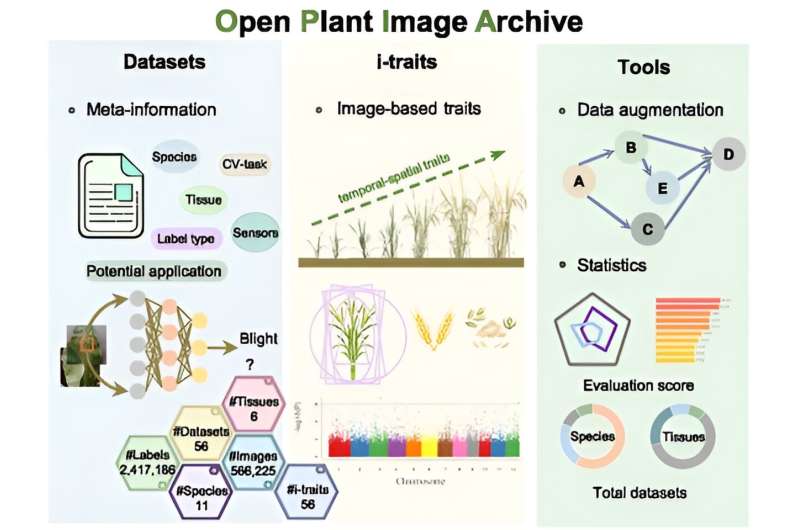
The OPIA team has integrated 56 high-quality plant image datasets covering 11 species and six tissue types with 566,225 images and 2,417,186 annotated instances using a standardized manual curation process. Notably, it incorporates 56 i-traits of 93 rice and 105 wheat cultivars based on 18,644 individual RGB images, and these i-traits are further annotated based on the Plant Phenotype and Trait Ontology (PPTO) and cross-linked with GWAS Atlas.
Additionally, each dataset in OPIA is assigned an evaluation score that takes account of the number of image samples, image quality, the richness of image samples, and the balance of image label categories, which provides users with an intuitive data quality evaluation. OPIA also provides tools for image preprocessing and intelligent prediction to assist with batch image data augmentation and preprocessing.
As a comprehensive resource archive of plant images and related traits, OPIA plays an important role in integrating the analysis of plant phenomics data from different acquisition platforms, tissue types, and phenotypic traits.
Through the application of image samples and corresponding label data from different sensor types, researchers are encouraged to further improve the accuracy of intelligent prediction methods, reveal the dynamic law of plant growth, and then promote the innovation and development of the global plant phenomics field.
More information: Yongrong Cao et al, OPIA: an open archive of plant images and related phenotypic traits, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad975
Journal information: Nucleic Acids Research , Nucleic Acid Research
Provided by Chinese Academy of Sciences