This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A novel machine learning model for molecular simulation under an external field
Prof. Jiang Bin's research team at the University of Science and Technology of China (USTC) have developed a universal field-induced recursively embedded atom neural network (FIREANN) model, which can accurately simulate system-field interactions with high efficiency. Their research was published in Nature Communications on October 12.
Atomic simulation plays a crucial role in understanding the spectra and dynamics of complex chemical, biological, and material systems at microscopic level. The key to atomic simulations is finding the accurate representation of high-dimensional potential energy surfaces (PESs).
In recent years, using atomistic machine learning (ML) models to accurately representing PESs has become a common practice. However, most ML models only describe isolated systems and can't capture the interactions between external fields and the systems, which can change the chemical structure and control the phase transition via field-induced electronic or spin polarization. A novel ML model taking external fields into account is urgently needed.
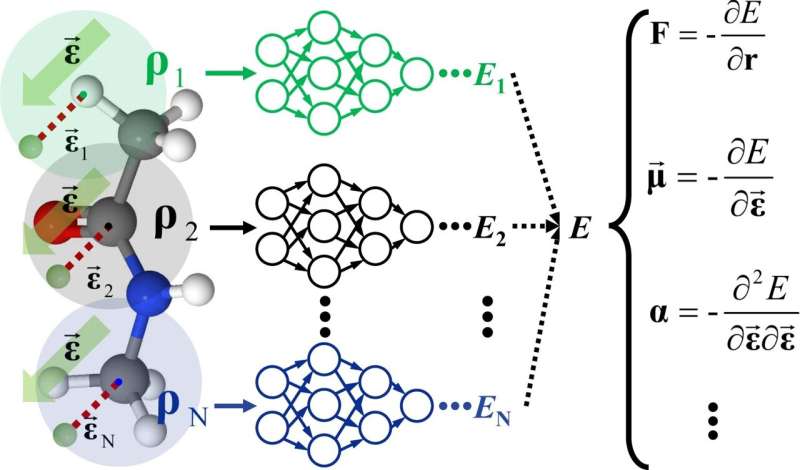
To tackle this problem, Prof. Jiang's research team proposed an "all-in-one" approach. The team first treated the external field as virtual atoms and used embedded atom densities (EADs) as descriptors to atomic environment. The field-induced EAD (FI-EAD) was derived from the linear combination of the field-dependent orbital and coordinate-based orbitals of atoms, which captures the nature of the interaction between the external field and the system, leading to the development of FIREANN model.
This model accurately correlates various response properties of the system, such as dipole moment and polarizability, with the potential energy changes that depend on external fields, providing an accurate and efficient tool for the spectroscopy and dynamics simulations of complex systems under external fields.
The team verified the capability of FIREANN model by conducting dynamical simulations of N-methylacetamide and liquid water under a strong external electric field, both demonstrating high accuracy and efficiency. It is worth mentioning that for periodic systems, FIREANN model can overcome the inherent multiple-value issue of the polarization by training with atomic forces data only.
This research filled the gap of lacking accurate external field representation in an ML model, which will contribute to the advancement of molecular simulations in chemistry, biology, and materials science.
More information: Yaolong Zhang et al, Universal machine learning for the response of atomistic systems to external fields, Nature Communications (2023). DOI: 10.1038/s41467-023-42148-y
Journal information: Nature Communications
Provided by Chinese Academy of Sciences