This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study finds novel gene evolution in the decaploid pitcher plant Nepenthes gracilis
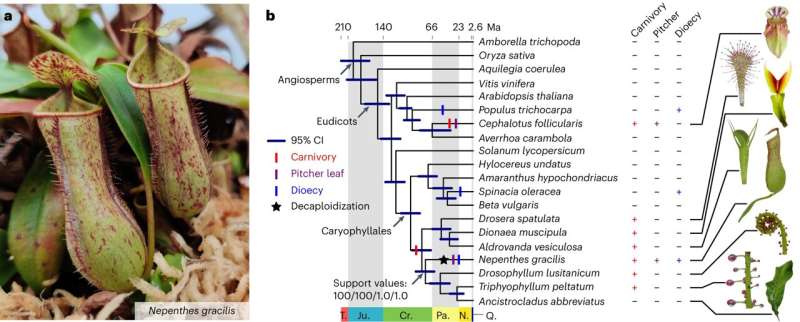
In a recent study, a team led by Würzburg botanist Kenji Fukushima investigated the genomic structure of the carnivorous pitcher plant Nepenthes gracilis and showed how polyploidy—the phenomenon of having more than two sets of chromosomes in cells—contributes to evolutionary innovation. Fukushima heads a working group at the Chair of Botany I at Julius-Maximilians-Universität Würzburg (JMU).
The results of the study have now been published in the journal Nature Plants.
On the trail of subgenome dominance
Plant genomes are known for their complex structure, and the study of polyploid genomes with multiple sets of chromosomes enables researchers to answer profound questions about genetics. "Our findings not only provide key insights into the adaptive landscape of the Nepenthes genome, but also broaden our understanding of how polyploidy can stimulate the evolution of new functions," states Professor Victor Albert from the University at Buffalo. Albert is the co-senior author of the study.
A central question deals with the mechanism of so-called subgenome dominance, which influences the retention and expression of genes across multiple chromosome sets.
This process often leads to a dominant subgenome, which is richer in genes. Recessive subgenomes, on the other hand, lose genes. In their study, Fukushima and his team focused on the previously unknown genomic structure of Nepenthes gracilis and investigated the complex relationship between the subgenomes.
Recessive subgenomes as innovative driving forces
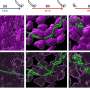
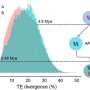
Using modern high-throughput sequencing techniques and bioinformatic analyses, the researchers were able to identify a decaploid genome structure—the fact that there are 10 sets of chromosomes in one cell—in the tropical pitcher plant. In doing so, they came across a clear signature of subgenome dominance, which led to one dominant and four recessive subgenomes.
"Interestingly, however, it turned out that it is the recessive subgenomes that are enriched with novel genes. This surprising discovery suggests that the recessive subgenomes make an important contribution to the evolutionary adaptation of the plant," says Fukushima. This is particularly true for defining traits such as pitcher leaves, which are used to catch insects, and dioecy, the presence of separate male and female plants.
In further analyses, the team identified specific genes on the recessive subgenomes that can be linked to these unique traits. One interesting example is a male-specific gene on the newly identified Y chromosome, which could play a crucial role in dioecy. They also discovered a cluster of genes specifically expressed in trapping pitchers that likely contributes to the evolution of carnivory in Nepenthes.
Possible applications in agriculture
In addition to researching the genetics of Nepenthes gracilis, the study contributes to the general understanding of the role of polyploidy in evolution and how novel genes arise. The knowledge gained could also be applied in other areas of plant research and improve our understanding of plant genomes and how plants can form complex and novel traits. Furthermore, the results allow conclusions to be drawn about plant diversity and adaptation in general and can make valuable contributions to agriculture, for example.
"Mechanisms of nutrient transport and reproduction are of particular interest there," explains Fukushima. The findings from the study could help to better understand these processes and thus contribute to sustainable and efficient agricultural practices.
More information: Franziska Saul et al, Subgenome dominance shapes novel gene evolution in the decaploid pitcher plant Nepenthes gracilis, Nature Plants (2023). DOI: 10.1038/s41477-023-01562-2
Journal information: Nature Plants
Provided by Julius-Maximilians-Universität Würzburg