November 27, 2023 feature
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
CRISPR-powered optothermal nanotweezers
Optothermal nanotweezers are an innovative optical design method that has revolutionized classical optical techniques to capture a broad range of nanoparticles. While the optothermal temperature field can be employed for in situ regulation of nanoparticles, challenges remain in identifying their potential for regulating bionanoparticles.
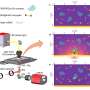
To observe the synergistic effects of optothermal manipulation and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based biodetection, the researchers developed a combination of CRISPR-powered optothermal nanotweezers abbreviated as CRONT.
In a new report in Light: Science & Applications, Jiajie Chen and a research team in optoelectronics engineering, biomedical engineering, and physics, accomplished this by harnessing diffusiophoresis and thermo-osmotic flows for optothermal excitation by successfully enriching DNA functionalized gold nanoparticles, CRISPR-associated proteins, and DNA strands.
The scientists built on an optothermal scheme to enhance CRISPR-associated single-nucleotide polymorphism detection at the single molecule level, to introduce a new CRISPR-based method to observe nucleotide cleavage. The researchers studied this innovative approach as a universal point-of-care diagnostics, biophotonics, and bionanotechnology field.
Optical tweezers
In 1986, Arthur Ashkin invented optical tweezers to regulate nano-objects remotely and received a Nobel prize in physics in 2018 for this groundbreaking discovery and contribution to biological systems. While classical optical tweezers depend on the momentum transformation of light, interdisciplinary combinations across plasmonic optics, electrical field and temperature have effectively addressed the limits.
A variety of innovative approaches have emerged to offer new opportunities in particle analysis and regulation. Optothermal nanotweezers use optical-induced thermodynamic forces to regulate nanoparticles in the micron-scale at sub-micron precision.
When compared to traditional optical tweezers, optothermal tweezers require a lower power density, making them an attractive alternative for biological detection, while reducing adverse optical effects on biological samples. Since thermal effects play a key role during a variety of biological processes, it is possible to leverage the capabilities of the temperature field for practical applications.
The method can be used to regulate bionanoparticles ranging from the micro-to-nanoscale to include bacteria and live cells, as well as single- and double-stranded DNA molecules and proteins.
Combining CRISPR with nanotweezers—CRONT
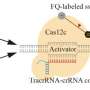
The clustered regularly interspaced short palindrome repeat (CRISPR) system itself offers a remarkable gene editing tool, which too received a Nobel prize in 2020. The method comprised a CRISPR-associated nuclease protein and a target DNA-specific guide RNA.
Biophysicists and bioengineers are increasingly keen to enhance the sensitivity and versatility of DNA detection by combining the CRISPR-Cas system with new sensing modes.
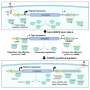
To overcome the existing limits of the method, Chen and colleagues designed a universally applicable optothermal tweezing platform known as CRISPR-powered optothermal nanotweezers to identify bionanoparticles and used the setup to identify in situ DNA molecules, without nucleic acid amplification. The experiments provided ultralow detection volumes at 10 μL to identify single nucleotide polymorphisms to study genetic diversity, disease susceptibility and drug response, to meet the future demands of genomic research and medicine.
The working principle
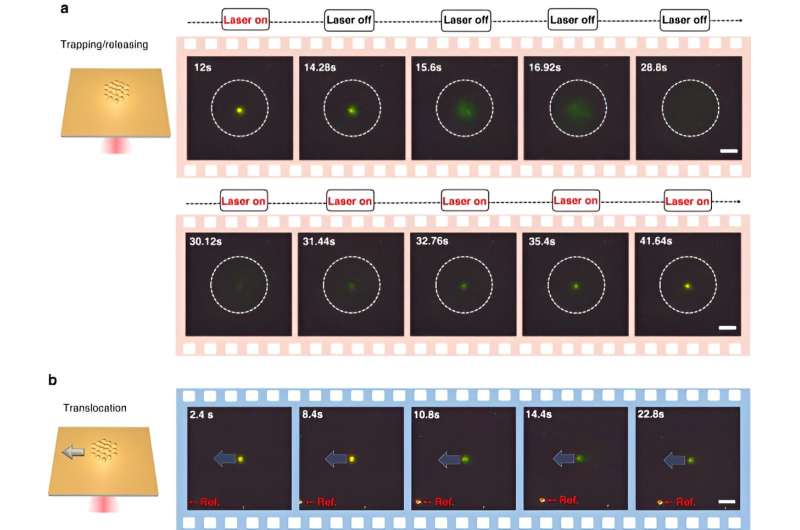
To enable CRONT (CRISPR-powered optothermal nanotweezers), the scientists designed a microfluidic chamber with a thin layer of gold film deposited on the cover glass. When the team irradiated the gold film with laser illumination, they generated a temperature field surrounding the laser spot. The scientists detailed optimal conditions of CRISPR reactions and initiated the cleavage of the DNA-gold nanofilm conjugate, using dark-field microscopy.
They added a nonionic polymer of polyethylene glycol (PEG) into the water solution as a biological surfactant for excellent biocompatibility.
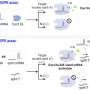
The presence of multiple nanoparticles and their varying thermophoretic mobility generated a distinct solute concentration. When solutes with increased concentrations influenced those with lower concentrations through osmotic pressure, the outcomes resulted in an interaction known as the diffusiophoretic force. This systematic investigation highlighted the potential for CRONT to be included to conduct biomolecular identification.
Optothermally combining proteins and DNAs
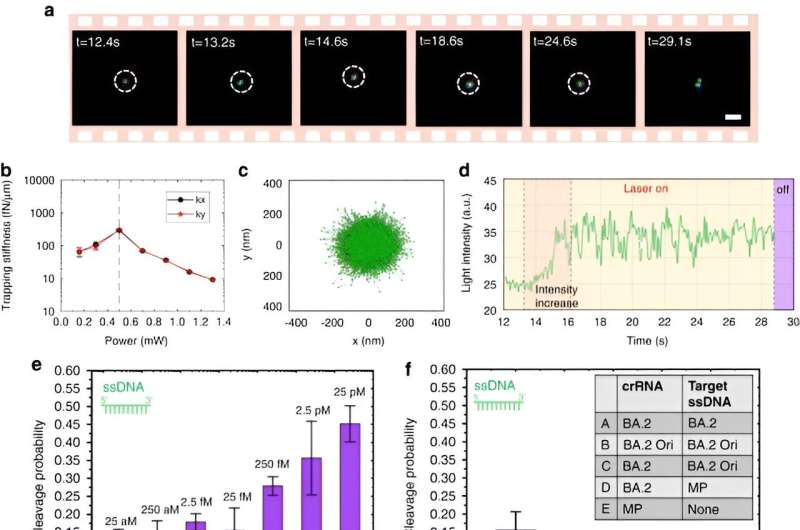
To enable CRISPR-powered optothermal nanotweezers, Chen and colleagues studied the aggregation behaviors of proteins and DNAs by using fluorescence labeling where the length of the rigid stem generated a polyethylene glycol concentration gradient. While a higher laser power did not continuously increase the accumulation rate due to an enlarged thermo-osmotic flow, the accumulation of single-stranded DNA was higher than double-stranded DNA. While protein accumulations are rarely studied in biophysics, the fluorescence-labeled Cas12a proteins showed a tendency to form slight ring-like accumulations, where increasing the laser power increased their accumulation rate.
The team additionally performed experiments on commonly incorporated proteins such as bovine serum albumin with FITC labeling. In the presence of an optothermal field, this protein distribution remained random and unaffected by the presence of polyethylene glycol molecules.
CRISPR-powered optothermal nanotweezers (CRONT) to identify nucleotides
Chen and team noted how the optothermal field associated with the CRISPR-powered optothermal nanotweezers (CRONT), provided a suitable temperature for CRISPR-based biodetection, with the capacity to enrich bionanoparticles to detect DNA at ultralow concentrations, instead of Brownian motion alone that is governed through the detection of diffusion.
The scientists included the CRISPR-12a scheme to examine single-stranded ambient DNA. The CRONT system successfully identified DNAs at the single molecule level for single nucleotide polymorphisms with high sensitivity and specificity.
Outlook
In this way, Jiajie Chen and colleagues incorporated diffusiophoresis and thermo-osmotic flows in the boundary layer of an optothermal responsive film to show a new method to regulate CRISPR-powered optothermal nanotweezers at the nanoscale.
This method allowed the immediate implementation of CRISPR-based biosensing with an ultralow detection volume.
Optical tweezers are endowed with DNA identification through CRISPR-based biosensing systems as an avenue for biomolecule enrichment to cleave the CRISPR complex. Such CRISPR-powered optothermal nanotweezers or CRONT systems hold immense promise to advance the understanding of complex biological processes as a versatile detection probe across biomedical research, drug discovery, and disease diagnostics.
More information: Jiajie Chen et al, CRISPR-powered optothermal nanotweezers: Diverse bio-nanoparticle manipulation and single nucleotide identification, Light: Science & Applications (2023). DOI: 10.1038/s41377-023-01326-9
Journal information: Light: Science & Applications
© 2023 Science X Network