This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New antimicrobial molecule shuts down bacterial growth without harming human cells
Scientists have shown how a molecule with broad-spectrum antibiotic activity works by disabling a process vital to bacterial growth without affecting the normal functioning of human cells. The journal mBio published the work, led by researchers at Emory University and Pennsylvania State University.
The molecule, known as KKL-55, is one of a suite of recently identified molecules that interfere with a key bacterial mechanism known as trans-translation, essentially shutting down the ability of bacteria to grow.
"We're opening a promising pathway for the development of new antibiotics to treat drug-resistant infections," says Christine Dunham, co-corresponding author of the paper and a professor in Emory's Department of Chemistry and the Emory Antibiotic Resistance Center.
Kenneth Keiler, a professor in the Department of Biochemistry and Molecular Biology at Pennsylvania State, is co-corresponding author of the paper. First authors are Ha An Nguyen, who did the work as an Emory chemistry Ph.D. candidate and has since graduated and works at Memorial Sloan Kettering, and Neeraja Marathe, a graduate student at Pennsylvania State.
A growing global threat
Antimicrobial-resistant infections have long been a public health threat. The situation grew even worse during the COVID-19 pandemic with increased antibiotic use and less prevention actions, according to the U.S. Centers for Disease Control and Prevention (CDC).
The CDC estimates that at least 2.8 million antimicrobial-resistant infections continue to occur in the United States each year, killing more than 35,000 people. Globally, the World Health Organization projects that these infections will cause up to 10 million deaths annually by 2050 if new antibiotics are not developed.
While antibiotics can save lives, any time they are used they can also contribute to the problem of resistance. Bacteria keep evolving new weapons as a defense against drugs, even as scientists work on developing new strategies to disarm bacteria.
Cross-toxicity, or harmful effects on humans, is another key drawback of some of the drugs used in a last-ditch effort to kill antibiotic-resistant bacteria.
Avoiding cross-toxicity
Dunham and Keiler are avoiding the problem of cross-toxicity by focusing on the inhibition of a mechanism unique to bacteria—trans-translation. This mechanism is vital to the proper functioning of the bacterial ribosome.
Keiler, a molecular geneticist and biochemist, first identified trans-translation in bacteria and is an expert in how it functions. Dunham, a structural biologist, is an expert in the human ribosome. She uses advanced biochemistry and structural biology techniques to understand the mechanics of molecular interactions.
"Our individual areas of expertise mesh well for this project," Dunham says. "By collaborating, we are able to take the science further, faster."
A cellular protein factory
The ribosome is an elaborate macromolecular machine within a cell that operates like a factory to manufacture proteins. Proteins are the machines that make cells run while nucleic acids such as DNA and RNA store the blueprints for life. The ribosome is made mostly of RNA, which does not just store information but can also act as an enzyme, catalyzing chemical reactions.
In a human cell, messenger RNA (mRNA), containing the instructions for manufacturing a protein, originates in the nucleus. While still in the nucleus, mRNA undergoes an elaborate quality-control process. It must pass inspection before getting exported to translate the information it contains into a protein.
"A lot of mRNAs have defects," Dunham says. "Human cells have efficient ways to test mRNAs and ultimately remove the defective ones."
Bacterial cells, however, have no nucleus or organized center for quality control.
"Bacteria wants to grow, grow and grow, which requires the ribosome to make a lot of proteins," Dunham says. "But when mRNA has defects, there is little to no quality control. When the ribosome encounters a defective mRNA protein, synthesis gets stalled."
The trans-translation process "rescues" ribosomes stalled due to such defects, in order to maintain proper protein synthesis and cell viability in bacteria.
How KKL-55 works
Using a high-throughput screening process, the Keiler lab has identified dozens of molecules that inhibit trans-translation in bacteria.
For the current paper, the researchers focused on understanding how one of these molecules, KKL-55, performs this trick. They used the high-powered structural biology technique of X-ray crystallography to capture KKL-55 in action as it interacted with a protein required for translation.
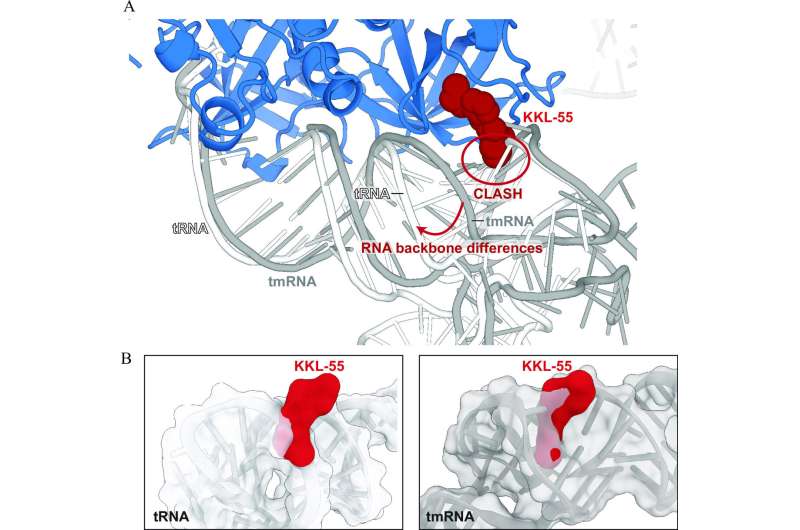
The results showed how KKL-55 blocks trans-translation by binding to elongation factor thermos-unstable (EF-Tu). EF-Tu is a protein that interacts with transfer RNA molecules, which play a key role in protein synthesis, and also transfer-messenger RNA, an RNA molecule required for the trans-translation pathway.
"We got lucky," Dunham says. "There are dozens of steps involved in the process that KKL-55 could have inhibited and we might have had to test for each one. But the results are clear-cut. It shuts down trans-translation right at the beginning by preventing EF-Tu from binding to tmRNA."
Determining the mechanism by which a molecule works to inhibit bacteria is a critical step toward developing a new antibiotic for clinical use. The next step is to test the efficacy of KKL-55 to treat a bacterial infection in a mouse model.
In 2021, the research team published their finding that a group of trans-translation inhibitors known as acylaminooxadiazoles clear multiple-drug-resistant Neisseria gonorrhoeae infection in mice after a single oral dose. That work is now advancing to clinical trials.
Dozens more trans-translation inhibitors await the team's investigation. Each represents a potential new weapon to help humans stay on top in the arms race with drug-resistant bacteria.
More information: Neeraja Marathe et al, Antibiotic that inhibits trans -translation blocks binding of EF-Tu to tmRNA but not to tRNA, mBio (2023). DOI: 10.1128/mbio.01461-23
Journal information: mBio
Provided by Emory University