This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Unlocking the genetic potential of poplars: A new era of precision genome editing with CRISPR
The Populus genus, commonly known as poplars, cottonwoods, and aspens, consists of approximately 30 tree species native to the northern hemisphere. Because of their diverse usages in landscape, agriculture, bioenergy, and industry, Populus species have been the focus of many tree breeding and genetic improvement programs.
Modern biotechnologies, including both genomics and genetic engineering (GE), have been considered as versatile tools for accelerating Populus domestication. Furthermore, since the P. trichocarpa genome sequence was released in 2006, followed by advancements in next-generation sequencing (NGS), researchers have been able to explore the genetic basis of crucial characteristics in Populus.
The CRISPR/Cas9 genome-editing tool has been widely utilized for creating knockouts in the hybrid poplar clone "717-1B4" (P. tremula x P. alba INRA 717-1B4). The development of alternative CRISPR-related tools, such as CRISRPa and base editing, has demonstrated great potential for improving plant fitness. However, the application of these new CRISPR tools in Populus remains very limited.
In May 2023, Horticulture Research published the research paper entitled "CRISPR/Cas9-based gene activation and base editing in Populus".
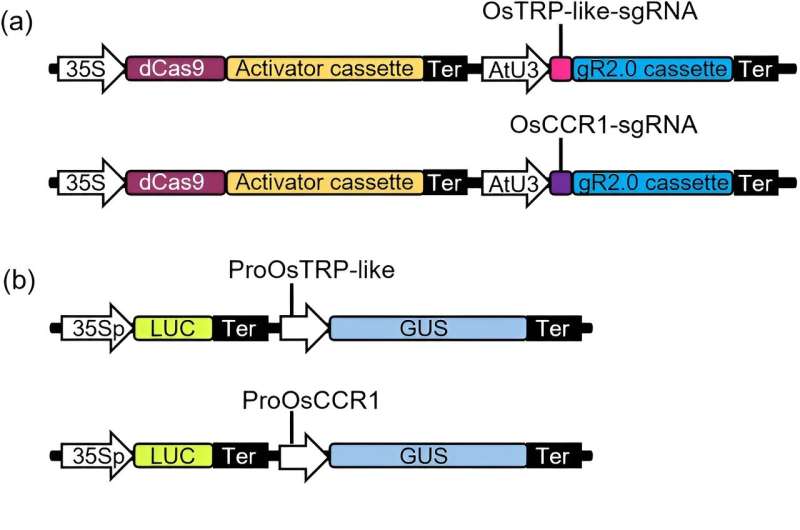
First, researchers successfully built a high-throughput reporter gene-based CRISPRa evaluation system in protoplasts. They cloned the promoters of OsTRP-like or OsCCR1 genes to generate reporter construct.
By co-transfection of OsTRP-like activation vector and its reporter vector in Arabidopsis protoplasts, the ratio of GUS/LUC is six-fold higher than those without the activation vector. Similar results were also acquired for OsCCR1 gene. They utilized this evaluation system to screen of sgRNAs for gene activation in Populus protoplasts.
TPX2 and LecRLK-G are crucial for plant growth and defense. They cloned 500-bp of promoter regions of TPX2 and LecRLK-G and inserted them into a reporter construct. Six and three sgRNAs were selected for TPX2 and LecRLK-G genes, respectively. By co-transfecting the reporter constructs and the activation construct containing each sgRNA, the activation efficiency of each sgRNA was assessed by GUS enzyme activities. Both the sgRNA6 of TPX2 and the sgRNA6 of LecRLK-G were selected for further stable transformation studies.
Stably transgenic poplars were then established using the identified sgRNAs. In hybrid poplar clone "717-1B4," the TPX2 gene's expression increased between 1.5-fold to 2.9-fold. Similarly, in clone "WV94," the LecRLK-G gene's expression escalated by 2-fold and 7-fold in different events.
These results demonstrate that the CRISPR-Act3.0 system is capable of activating endogenous TPX2 or LecRLK-G genes in stable transgenic poplars. Further, the research aimed to truncate the PLATZ protein, linked to disease-related genes, by introducing a premature termination codon using the Cas9 nickase (nCas9)-based cytosine base editor (CBE) in poplar clone "717-1B4."
After identifying two potential sites for introducing stop codons, the researchers tested two base editors (pHEE901(BE3) and A3A/Y130F-BE3) in poplar protoplasts. Both editors effectively induced C-to-T mutations in the protoplast system. In addtion, pHEE901(BE3) also showed expected base editing in stable transgenic poplars.
In summary, this study introduces a novel application of CRISPR tools, specifically CRISPRa and base editing, to enhance gene function in two Populus species. Remarkably, the dCas9-based CRISPRa system is shown to effectively activate genes in different Populus genotypes, with potential implications for phenotypic changes in future investigations.
The research establishes a vital foundation for genetic improvements in Populus, showcasing the expansive potential of CRISPRa and base editing in woody species, promising advancements in tree breeding and other horticultural plants.
More information: Tao Yao et al, CRISPR/Cas9-based gene activation and base editing in Populus, Horticulture Research (2023). DOI: 10.1093/hr/uhad085
Journal information: Horticulture Research
Provided by NanJing Agricultural University