This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
New tools for fungicide resistance detection

Researchers at the Center for Crop and Disease Management (CCDM) have developed a new method for detecting fungicide resistance, enabling them to detect multiple mutations, both known and novel, in just one test.
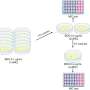
With co-investment by Curtin University and GRDC, CCDM researchers were able to rapidly and accurately detect fungicide resistance mutations using a portable DNA sequencing device called the MinION from Oxford Nanopore Technologies in the United Kingdom, including unknown mutations that could fail to be picked up by traditional methods.
The research was published in Scientific Reports.
CCDM researcher and lead author Dr. Katherine Zulak said traditional techniques for identifying mutations within pathogens often involve labor-intensive and time-consuming processes and are limited to screening for only previously known mutations.
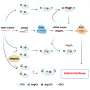
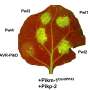
"Barley net blotch has two forms, spot form and net form net blotch, each with its own suite of different mutations," Dr. Zulak said.
"Furthermore, the particular target gene we were looking at can have mutations occur in two different regions, both of which again have their own suite of mutations.
"So, every time we need to investigate a sample for fungicide resistance, we previously had to run tests for every one of these possible mutations.
"To simplify this increasingly complicated process, we've used the Nanopore MinION to sequence fungicide target genes and provide a comprehensive map of all possible mutations, which may include those that we hadn't previously identified.
"For example, while developing this method, we found a recently discovered variation of a known mutation. This is the type of mutation that could go unnoticed with traditional methods."
Dr. Zulak said that further research and development is required before field detection of fungicide resistance becomes a reality. However, the data from the sequenced target genes could contribute to a national resource that can be continuously utilized by future researchers.
"The fungicide resistance landscape is always changing, and this technology simplifies our fungicide resistance detection process and ensures we have the capacity to detect new mutations as they appear, putting us in a better position to tackle this issue as it evolves.
"As we sequence regions within the pathogen's genome, we are not only gathering data for current research but also creating a resource for future studies. This genetic information can be stored in national databases, providing a reference point for researchers in subsequent projects and ensuring that future efforts build on our existing knowledge."

Professor Mark Gibberd, CCDM director, highlighted the center's commitment to seeking new tools and resources which can expedite and enhance the industry's ability to combat this problem.
"Innovation lies at the core of our research center, and this project stands as a great example of our ability to draw inspiration from diverse fields to confront the most significant disease challenges in the grains industry," Professor Gibberd said.
"Research investment in Australia is only a small fraction of global research investment. Through CCDM's international partnerships, co-investments, and collaborations, we've been able to leverage the outputs of global R&D to bring world-class innovations and solutions to benefit Australian agriculture."
More information: Katherine G. Zulak et al, Exploiting long read sequencing to detect azole fungicide resistance mutations in Pyrenophora teres using unique molecular identifiers, Scientific Reports (2024). DOI: 10.1038/s41598-024-56801-z
Journal information: Scientific Reports
Provided by Grains Research and Development Corporation