This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers unveil comprehensive database of published microbial signatures
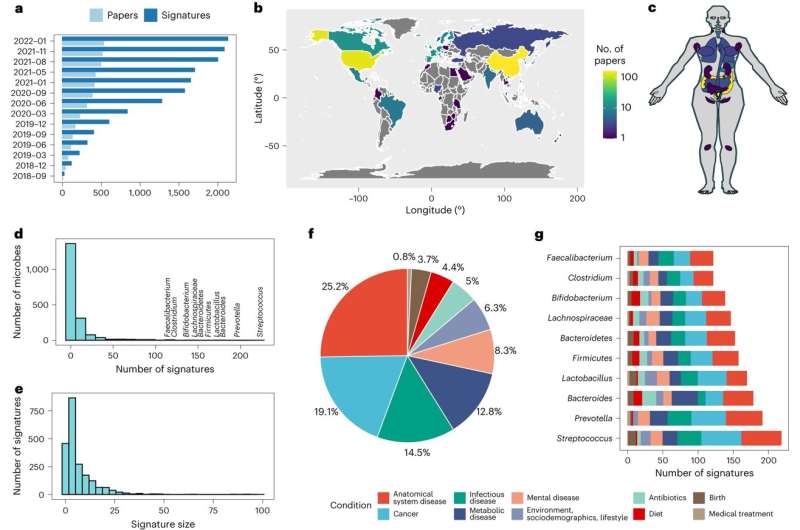
A new study published by researchers from the CUNY Institute for Implementation Science in Population Health (ISPH) at CUNY SPH and colleagues presents BugSigDB, a community-editable database of manually curated microbial signatures from published studies.
The paper titled "BugSigDB captures patterns of differential abundance across a broad range of host-associated microbial signatures" is available in Nature Biotechnology, and the public wiki is available at https://bugsigdb.org.
The database records essential methods and results to enable high-throughput analysis of similarity of microbial signatures identified by independent studies, of co-occurrence and co-exclusion of individual microbes and of consensus signatures conserved across multiple studies of similar health outcomes and exposures. It allows assessment of microbiome differential abundance within and across experimental conditions, environments or body sites.
First author Ludwig Geistlinger started the project as a postdoctoral student at CUNY SPH. He is now associate director of computational biology at the Center for Computational Biomedicine at Harvard Medical School.
"BugSigDB is the first comprehensive collection of published microbial signatures that can be used to compare host-associated differential microbial abundance across independent studies," says Dr. Geistlinger. "It helped us to uncover reproducible patterns of differential microbial abundance within and across health outcomes that we couldn't notice from just reading the published literature without standardizing it."
"Having the opportunity to work with and mentor the BugSigDB interns—many of whom were CUNY SPH students—has been truly amazing," says recent CUNY SPH doctoral graduate Chloe Mirzayi, the study's second author. "Since the project started, I have gotten to see these bright and motivated students contribute to BugSigDB and grow as researchers as they have developed skills in critically reading literature, interpreting study results and performing secondary data analysis."
"This is the most significant project I've ever undertaken," says Professor Levi Waldron, the study's senior author. "It's the product of four years of work by nearly 60 student curators who entered nearly 3,000 microbial signatures from 750 studies and supported by multiple software developers, CUNY students and collaborators who helped manage teams of curators to develop novel methods to learn from this new type of database.
"BugSigDB is powered by the same technology as Wikipedia, so my next goal is to recruit more editors and ensure this becomes a living database maintained by the whole microbiome research community."
More information: Ludwig Geistlinger et al, BugSigDB captures patterns of differential abundance across a broad range of host-associated microbial signatures, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01872-y
BugSigDB—a database for identifying unusual abundance patterns in human microbiome studies, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01930-5
Journal information: Nature Biotechnology
Provided by The City University of New York