This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop first method to study microRNA activity in single cells
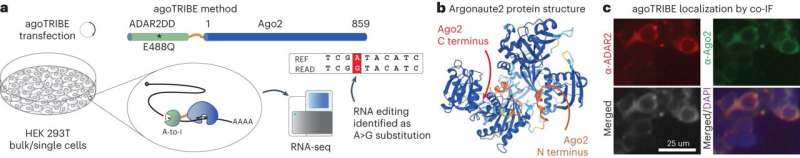
MicroRNAs are small molecules that regulate gene activity by binding to and destroying RNAs produced by the genes. More than 60% of all human genes are estimated to be regulated by microRNAs; therefore, it is not surprising that these small molecules are involved in many biological processes including diseases such as cancer.
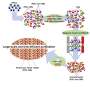
To discover the function of a microRNA, it is necessary to find out exactly which RNAs are targeted by it. While such methods exist, they require a lot of material typically on the order of millions of cells, to work.
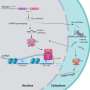
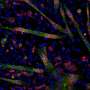
Now researchers at Stockholm University and SciLifeLab have developed a new method to detect microRNA targets at the level of single cells. Such cells are each around one-hundredth millimeter in diameter and weigh less than a billionth gram, and comprise the basic building blocks of living organisms.
With their new sensitive method, the researchers can follow microRNA targeting of thousands of RNAs during biological processes such as the cell cycle or differentiation into red blood cells. In these processes, the researchers find that microRNAs—surprisingly—perform quite different tasks in each cell. In the future, it will be possible to also apply this method to study microRNA targeting in whole tissues, to find out exactly what is happening in each of the many cell types that comprise complex organs such as brains.
Marc Friedländer, associate professor at Stockholm University, says, "In our research team, we want to understand and ultimately make mathematical models of gene regulation at the level of the single cell. Our new method is a huge leap towards making this possible."
The work was spearheaded by Dr. Inna Biryukova, who took a leading role in developing the laboratory method, and by Ph.D. student Vaishnovi Sekar, who performed the bulk of the advanced computational analyses.
Vaishnovi Sekar says, "In terms of complexity of the computational work, this is uncharted territory, and we lacked reference points and thresholds. We had to explore a myriad of approaches to devise a methodology that not only works but also yields biologically meaningful observations."
The study has been published in the journal Nature Biotechnology.
More information: Inna Biryukova et al, Detection of transcriptome-wide microRNA–target interactions in single cells with agoTRIBE, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01951-0. www.nature.com/articles/s41587-023-01951-0
Journal information: Nature Biotechnology
Provided by Stockholm University