This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
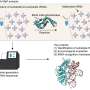
Mechanism of methyltransferase METTL8-mediated mitochondrial RNA m3C modification and its relaxed substrate specificity
A study published in the journal Science Bulletin was led by Profs. Xiao-Long Zhou and En-Duo Wang (CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences).
tRNA is a key adaptor molecule in mRNA translation. There are a large number of post-transcriptional modifications on tRNA, which regulate the speed and fidelity of protein synthesis. 3-Methylcytosine (m3C) modification is widely found at position 32 (m3C32) of the anticodon loops of several cytoplasmic and mitochondrial tRNAs in eukaryotes.
A previous study by the same lab has found that the m3C32 modification of human cytoplasmic tRNAs was mediated by METTL2A/2B and METTL6, while that of human mitochondrial tRNAThr (hmtRNAThr) and tRNASer(UCN) (hmtRNASer(UCN)) is catalyzed by METTL8; Human METTL8 generates two protein isoforms of different lengths by alternative splicing of mRNA.
The long-length form, METTL8-Iso1, was targeted into mitochondria to catalyze the m3C32 modification of hmtRNAThr and hmtRNASer(UCN); while the short-length form, METTL8-Iso4, was located in the nucleolus with unknown function.
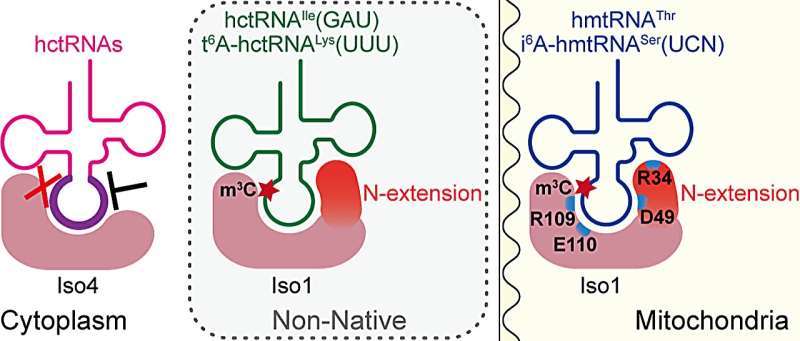
The only difference between the two isoforms is a 28-amino acid N-terminal extension peptide in METTL8-Iso1. Whether METTL8-Iso4 has m3C32 methyltransferase activity and the role of the N-terminal extension of METTL8-Iso1 in mitochondrial tRNA m3C32 modification is unknown.
It is also unclear whether cytoplasmic or mitochondrial m3C32 modification enzymes can cross-recognize tRNAs from different cellular compartments. In addition, since most tRNA m3C32 modifications require N6-threonylcarbamoyl adenosine modification at position 37 (t6A37) in the anticodon loop as a prerequisite, the preparation of tRNA molecules containing only m3C32 modification has not been fully achieved.
To address these questions, the researchers confirmed the conservation of the N-terminal extension (N-extension) of METTL8-Iso1 through multiple sequence alignment. In vitro enzyme activity determination revealed that METTL8-Iso4 had no m3C32 modification activity. They further proved that the N-extension of METTL8-Iso1 acted as a key tRNA-binding element in the catalytic process.
Two completely conserved amino acid residues in all METTL2A/2B/8 proteins were identified. METTL8-Iso1 was able to mediate m3C32 modification for both cytoplasmic and E. coli tRNAs, which was not reliant on t6A37.
However, cytoplasmic m3C32 modification enzymes METTL2A and METTL6 were unable to catalyze m3C32 modification of mitochondrial tRNA, indicating that METTL8-Iso1 has a more relaxed substrate specificity. The m3C32 modification did not affect the t6A37 modification and aminoacylation levels of hmtRNAThr.
Finally, they also revealed that METTL8-Iso1 interacted with mitochondrial seryl-tRNA synthetase (SARS2) and mitochondrial threonyl-tRNA synthetase (TARS2), respectively, and significantly promoted aminoacylation activity of SARS2 and TARS2.
In summary, this work reveals the molecular mechanism of mitochondrial tRNA m3C32 biogenesis mediated by METTL8, which relies on a specific N-extension as a key RNA-binding element. METTL8 had a broad spectrum of heterogenous tRNA substrates, which provided a basis for preparation of tRNAs containing only a m3C moiety. This work provides a comprehensive understanding of the conservation and difference between cytoplasmic and mitochondrial tRNA m3C modification.
More information: Meng-Han Huang et al, Mitochondrial RNA m3C methyltransferase METTL8 relies on an isoform-specific N-terminal extension and modifies multiple heterogenous tRNAs, Science Bulletin (2023). DOI: 10.1016/j.scib.2023.08.002
Provided by Science China Press