This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
How hydrophobicity shapes protein assemblies
Through a nuanced balance of electrical and hydrophobic forces, biological molecules self-assemble into the large functional structures that maintain life's vital functions. Understanding how proteins self-assemble requires knowledge of both forces. But while predicting the electrical interactions of individual proteins is simple, deriving their hydrophobic ones is less straightforward.
In a study published in The European Physical Journal E, Angel Mozo-Villarias, of the Autonomous University of Barcelona, Spain, and his colleagues develop a formulation for how proteins align into membrane-like structures based on hydrophobic interactions. The model could help to predict the configuration of macromolecular assemblies at any scale, providing a useful tool for novel materials and drug discovery research.
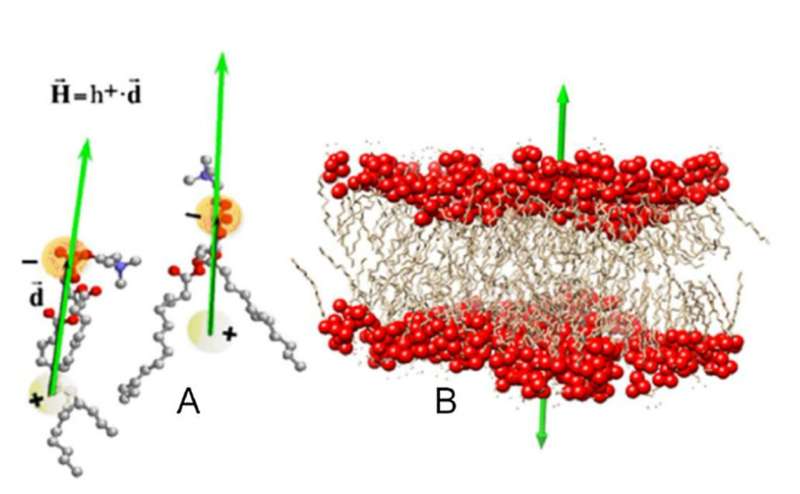
Hydrophobicity is an emergent property of complex molecular systems that does not manifest itself in each individual component. To examine hydrophobicity in proteins, researchers assign each of their constituent amino acids an index on a scale according to how much energy it takes to transfer them from a hydrophilic medium to a hydrophobic one. For a collection of amino acids, these indexes—the "hydrophobic charges"—establish a hydrophobic field much like a distribution of electrical charges establishes an electric field.
Based on the distribution of hydrophobic charges on a protein, Mozo-Villarias and his colleagues first defined a vector that described the protein's dipolar character. Then, using an electrical analogy, they calculated the energy stored in a system formed by two hydrophobic dipoles. Simulations showed that the hydrophobic dipoles tended to align parallel to each other, following the tendency of phospholipid molecules to orient themselves to form a double-layer in biological membranes.
This membrane effect provides a hydrophobic mechanism by which self-assembling proteins align before establishing more permanent bonds. The researchers conclude that effect is a general principle that gives rise to the great variety of morphologies in protein assemblies seen in nature.
More information: Juan A. Cedano et al, How hydrophobicity shapes the architecture of protein assemblies, The European Physical Journal E (2023). DOI: 10.1140/epje/s10189-023-00320-8
Journal information: European Physical Journal E
Provided by Springer