This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Exploring the OsMATL2 gene as a candidate for haploid induction in rice
Conventional crop breeding is not time-efficient for creating inbred lines with desired genetic traits owing to the diploid nature of plants, wherein they have two sets of chromosomes, one from each parent. In contrast, double haploid technology utilizes gene-edited haploid inducer plants to create double haploid crops that have two sets of chromosomes from a single parent. This revolutionary agricultural method can create inbred crop lines in a single generation thereby accelerating the breeding process.
Recent studies have unveiled the potential of specific genes in triggering haploid induction, a key step in double haploid technology. In particular, the gene ZmMATL was identified as a pollen-specific phospholipase in maize that plays a role in haploid induction within the plant's reproductive processes.
Building on this discovery, researchers demonstrated the conservation of pollen-specific phospholipase A-mediated in vivo haploid induction across various monocot species including indica rice by mutating the OsMATL gene. However, the haploid induction rate (HIR) was found to be only 6% at best, which is far below the industry standard, suggesting a need to increase this rate.
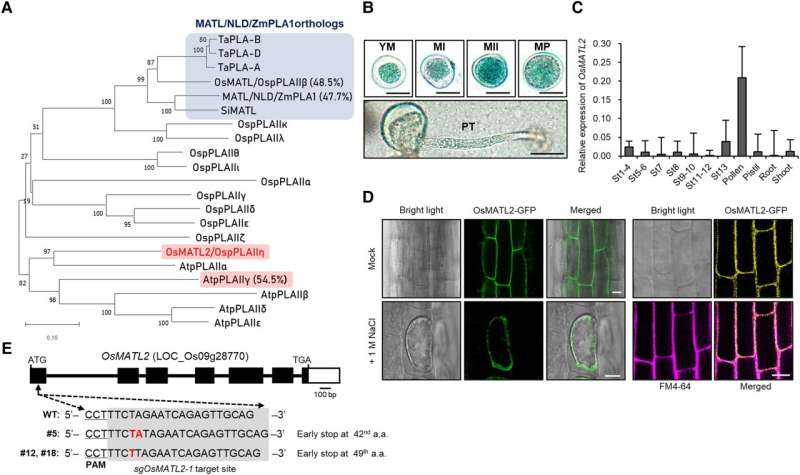
In a recent collaborative study, researchers from Korea led by Dr. Yu-Jin Kim from Pusan National University identified OsMATL2, a potential haploid-inducing gene in Japonica rice (Oryza sativa japonica). Their research was published in Plant Physiology.
Using a combination of GUS reporter genes, green fluorescent protein-tagged antibodies, and reverse transcriptase quantitative polymerase chain reaction, the team observed that OsMATL2 protein, a phospholipase enzyme, is highly expressed in pollen, primarily in the plasma membrane of cells of the japonica rice plant.
To evaluate the role of OsMATL2—the gene encoding OsMATL2 protein—in haploid induction, the team used the CRISPR/Cas9 system to generate knockout mutants (sgOsMATL2) with suppressed OsMATL2 protein expression.
While diploid sgOsMATL2 plants showed no vegetative defects compared to normal rice, their haploid counterparts were notably smaller in size. Additionally, all sgOsMATL2 plants showed reduced (around 80% of normal) seed setting. Most importantly, haploid sgOsMATL2 plants showed male sterility due to severe defects in pollen development.
Through flow cytometry and fluorescence microscopy, the researchers observed that haploid sgOsMATL2 plants possessed only half as many chromosomes as normal, confirming that inactivation of the OsMATL2 gene does indeed trigger haploid induction. The HIR of sgOsMATL2 plants was observed at 6.34% on average, which is slightly higher than the HIR triggered by OsMATL mutation.
"Traditional crop breeding requires multiple generations of self-crossing. Gene-edited OsMATL2 plants can be used as a haploid inducer to produce perfectly inbred rice lines in a single generation," Dr. Kim says.
This study showed the existence of a new haploid-inducing gene in rice. While OsMATL and OsMATL2 genes have an individual HIR of ~6%, with their functional redundancy, it may be possible to mutate both genes to obtain a higher HIR. The applications of this research can revolutionize rice cultivation.
"Rapid crop breeding is required to combat climate change, abiotic stress, and threats from viruses. Identification of more haploid-inducing genes and understanding their mechanisms during double fertilization in plants can reduce the time and effort needed for effective crop breeding," concludes Dr. Kim
More information: Jin Hoon Jang et al, Loss of function of pollen-expressed phospholipase OsMATL2 triggers haploid induction in japonica rice, Plant Physiology (2023). DOI: 10.1093/plphys/kiad422
Provided by Pusan National University