This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Study sheds light on agrobacterium-mediated transformation in key rice cultivars
Rice (Oryza sativa L.), the staple that feeds over half of the globe, has undergone significant transformation and regeneration advancements over the last two decades. Although agrobacterium-mediated transformation has been successful in several rice cultivars, it has encountered efficiency obstacles, particularly in newly developed indica cultivars from South China. The absence of a consistent 'gold standard' protocol for transformation, particularly in indica varieties, highlights the need for more in-depth exploration and understanding.
A paper, entitled "Agrobacterium-mediated transformation efficiency and grain phenotypes in six indica and japonica rice cultivars," was published in Seed Biology on 30 March 2023.
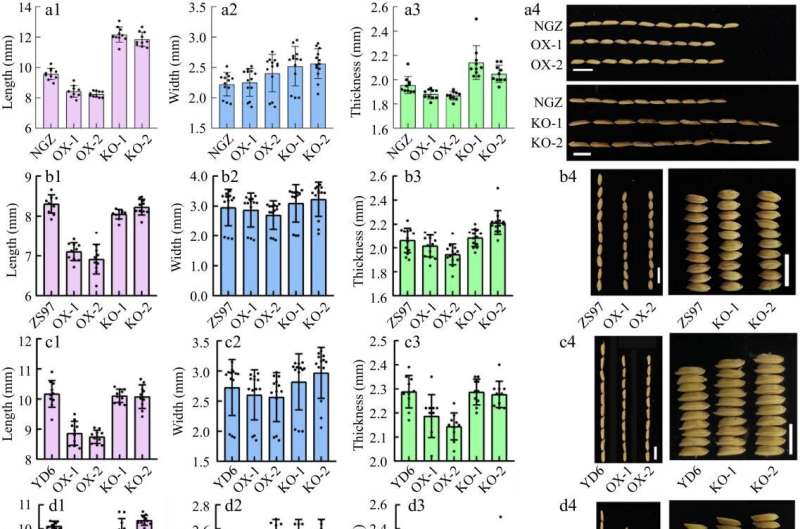
In this study, the tissue culture and transformation performance of six rice cultivars were evaluated, with a focus on the indica cultivar Nanguizhan (NGZ), which was previously developed for its temperature sensitivity.
Firstly, researchers compared the ZH11 cultivar, known for its efficiency in agrobacterium-mediated transformation, with five other indica rice cultivars that differed in grain-related agronomic traits, and they found that these cultivars were highly diverse in critical grain-related agronomic traits. NGZ was identified as a superior grain quality rice cultivar due to its low chalky grain rate and desirable grain attributes along with the shape and milled performance, especially favorable for grain size and shape functional studies.
To generate transgenic plants and evaluate the performance of callus culture, researchers used two plasmids: one for GSN1 overexpression and the other for knocking out GSN1 through CRISPR-Cas9 gene editing.
Based on phenotype and statistical analysis, as well as callus induction rate, the ranking of these six varieties is as follows: ZH11 (88.7%), ZS97 (83.3%), NGZ (82%), YD6 (77.6%), HHZ (72.7%) and Kasalath (64.2%).
These results confirmed that the indica rice cultivars NGZ has good callus induction ability under commercial Callus Induction Medium (CIM) conditions. Moreover, T2 seed phenotypes from the six cultivars confirmed successful transformations and phenotypic changes consistent with genetic modifications.
To further explore the molecular basis for the difference in transformation efficiency between indica and japonica rice, researchers validated the expression patterns, SNPs and InDels of selected genes related to callus regeneration among these cultivars. The research results indicated that genes related to callus induction might contribute to the browning and induction rate changes of callus in japonica and indica rice cultivars.
In summary, this study provides a deeper understanding of the agrobacterium-mediated transformation process specific to rice. The revelation about differential responses across cultivars, combined with the nuanced grain phenotype changes post-transformation, holds profound implications. It shows potential for improvement in transformation techniques and ultimately aiding the development of robust and high-yielding rice variants in the future, addressing global food security concerns.
More information: Ke Chen et al, Agrobacterium-mediated transformation efficiency and grain phenotypes in six indica and japonica rice cultivars, Seed Biology (2023). DOI: 10.48130/SeedBio-2023-0004
Provided by Maximum Academic Press




















