This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Population genomic analyses reveal multiple domestications of Asian rice
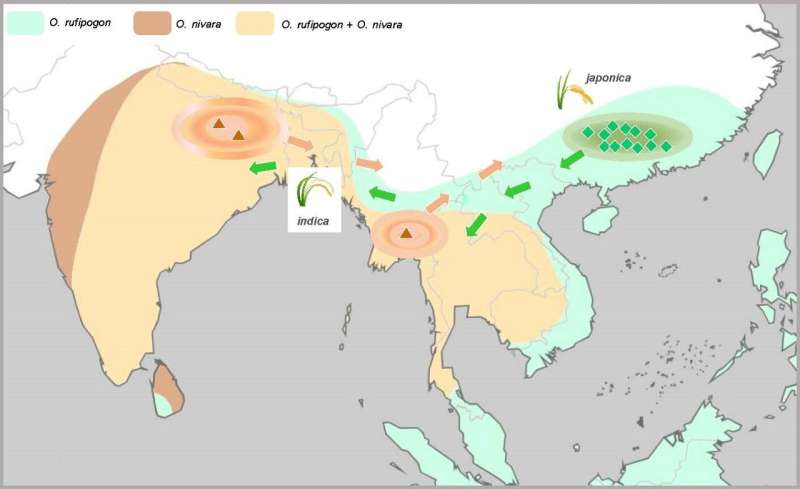
In a study published in Nature Plants, researchers led by Prof. Ge Song from the Institute of Botany of the Chinese Academy of Sciences have demonstrated that Asian rice domestication began independently from divergent wild lineages in different areas. It was then completed by continuous selection and the exchange of beneficial alleles between different cultivar groups.
Domesticated Asian rice (Oryza sativa L.), consisting of two subspecies (Japonica and Indica), is not only one of the world's most important crops but also an excellent model for biological research. Despite extensive study, the origin of the domestication of Asian rice has been controversial for almost a century. Of the various opinions, two leading hypotheses (single vs. multiple domestication) have been widely accepted, but the original controversy has remained unresolved.
In this study, the researchers carried out extensive analyses of the domestication history of rice at the genome level using a dataset of 459 newly re-sequenced and 1,119 publicly available genomes of wild and cultivated accessions.
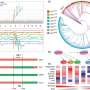
By proposing a new strategy for testing alternative hypotheses, they identified 993 selected genes that were shared by Japonica and Indica, suggesting that the domestication alleles of these genes originated only once in either Japonica or Indica.
Importantly, the researchers showed that the domestication alleles of most of the selected genes (~80%) originated from wild rice O. rufipogon in China. However, the domestication alleles of a substantial minority of the selected genes (~20%) originated from wild rice O. nivara in South and Southeast Asia. These results demonstrate separate domestication events for Asian rice in different areas.
In addition, this study contributed genomic resources, including 422 newly re-sequenced wild accessions verified by phenotypic examination, that will greatly facilitate further studies of rice genetics, evolution and breeding.
The expanded dataset allowed the researchers to uncover the deep genetic structure as well as multiple distinct lineages within both wild and domesticated rice, providing a better phylogenetic framework for inferring the evolutionary history of domestication genes.
Unexpectedly, the researchers found that some selective sweep regions in rice contained genes from different evolutionary origins. This discovery suggests that the previous practice of reconstructing a single phylogeny based on concatenating sweep regions to infer domestication history is unwarranted.
"This study has important implications for other domesticated species, especially those with confounded evolutionary histories defined by widespread and continuous gene flow among cultivars and lines," said Prof. Ge Song, corresponding author of the study.
More information: Chun-Yan Jing et al, Multiple domestications of Asian rice, Nature Plants (2023). DOI: 10.1038/s41477-023-01476-z
Journal information: Nature Plants
Provided by Chinese Academy of Sciences