This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers explore use of AI to improve COVID-19 drug design
Discovering and developing drugs can be slow, costly and high-risk, but harnessing advances in artificial intelligence (AI) can help with these processes, say Brock University researchers.
Yifeng Li, Canada Research Chair in Machine Learning for Biomedical Data Science, and his Computer Science master's student Cameron Andress published a paper, in PLOS Computational Biology, on how AI can better create a drug to prevent the SARS-CoV-2 virus—responsible for contracting COVID-19—from entering human cells compared to more conventional methods.
"This is a unique and important piece of work in the AI for drug design community," says Andress, who is the paper's first author. "Our results suggest that AI is capable of producing well-suited drug candidates for a chosen virus, which can significantly accelerate the drug development process and potentially save more lives."
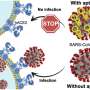
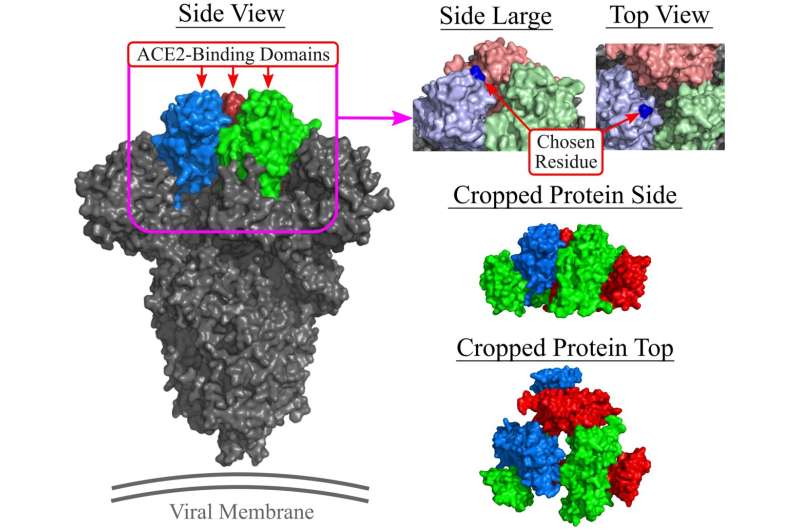
Li and his team focused on a substance called an aptamer, which can function as a drug. Aptamers are short, single-strand DNA and RNA molecules that bind strongly and exclusively to a particular protein that has been targeted to treat or prevent illness.
Aptamer drugs are relatively new and innovative, says the Assistant Professor of Computer Science. They are stronger and more precise than therapeutics known as "small molecule drugs"—such as Aspirin and penicillin—in which molecules can pass through membranes to reach the protein they are targeting.
Aptamers are usually developed through a series of experiments in which a group of random aptamers is added to a controlled environment containing the virus. Scientists measure how well the aptamer binds to the target protein and will keep making a series of modifications until the bond is strengthened.
Andress turned to machine learning and created a program that simulates the physical process by generating multitudes of different DNA and RNA sequences and testing how strongly and precisely they bind with the target protein.
"AI is a new paradigm for drug discovery as it dramatically reduces development time and cost and is able to efficiently search the vast molecular space," he says.
Li recalls how he and Andress became interested in creating aptamer drugs to treat COVID-19 in spring 2020 during the onset of the pandemic.
"We chose COVID-19 because of its urgent nature and long-lasting medical challenges," says Li. "Our work is among the first to design aptamer drugs for COVID."
Li says the team's model can be applied to address future health crises.
"For future similar COVID-19 variants or other pandemics caused by other types of viruses, since we have the framework, experience and a team ready, we can replace the current spike protein with the new target protein of interest," he says.
More information: Cameron Andress et al, DAPTEV: Deep aptamer evolutionary modelling for COVID-19 drug design, PLOS Computational Biology (2023). DOI: 10.1371/journal.pcbi.1010774
Journal information: PLoS Computational Biology
Provided by Brock University