This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New method brings increased efficiency, precision and reliability in DNA editing
In a new study published in Nature Methods, researchers at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany, describe improvements in the methods with which mutations can be introduced in human and other genomes—making these methods much more efficient and less error prone.
In the field of genome editing, scientists often need to change one letter—corresponding to one of the DNA bases Adenine, Guanine, Cytosine or Thymine—to another letter at one specific position in the genome. To do this, they use reagents that cut both strands of the DNA close to the position they want to change.
They then provide the cell with DNA molecules that contain the desired new letter in the hope that the cell's repair systems will use these molecules to introduce the desired mutation when the DNA break is repaired. Since different repair systems in the cells compete with each other and only one of these systems is able to introduce the desired new mutation, applications of genome editing of single letters have so far been limited by low efficiency and unintended byproducts.
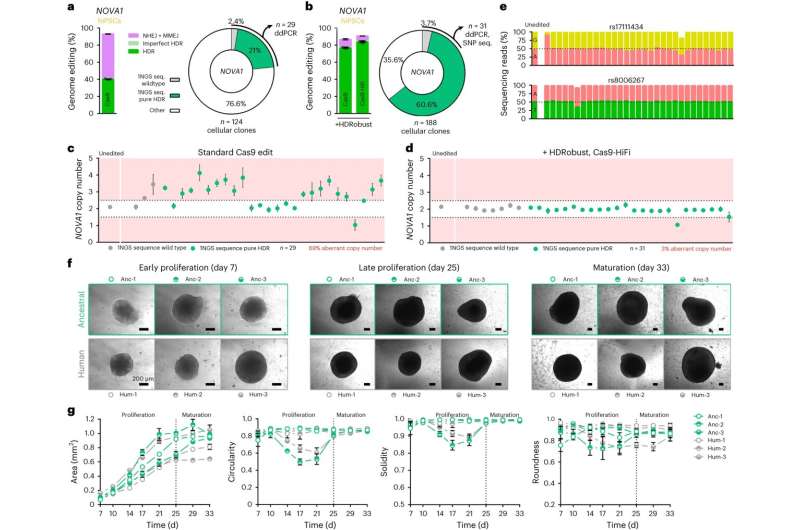
Through the combined inhibition of two repair pathways that compete with the desired one, the team achieved the induction of point mutations in up to 93 percent of chromosomes in populations of cells. Importantly, this method largely abolishes unwanted insertions, deletions, rearrangements in the area where the desired mutation is located as well as unintended changes at other sites in the genome. It therefore greatly increases the precision and reliability of the DNA editing process.
Studying the effects of mutations
The researchers demonstrated the efficiency of the novel method in the laboratory by changing 58 different target sites in human cells. This method is likely to have many applications in basic research. "We look forward to using this method to more easily introduce genetic changes into cells to study the effects of mutations that set modern humans apart from Neandertals," says Svante Pääbo, who helped facilitate the work.
The scientists also corrected pathogenic mutations in cells derived from patients suffering from three genetic diseases: anemia, sickle cell disease, and thrombophilia.
"The implications for helping cure human diseases in the future are potentially vast. One could imagine removing cells from patients, editing them with the help of this method, and then giving them back to the patients," says Stephan Riesenberg, who led the work. "Nevertheless, the road from demonstrating that the method works in the laboratory to applying it to patients is still a long one," he cautions.
More information: Stephan Riesenberg et al, Efficient high-precision homology-directed repair-dependent genome editing by HDRobust, Nature Methods (2023). DOI: 10.1038/s41592-023-01949-1 www.nature.com/articles/s41592-023-01949-1
Journal information: Nature Methods
Provided by Max Planck Society