This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Machine learning meets behavioral neuroscience: Allowing for a more precise phenotyping
A new computer program allows scientists to observe the behavior of multiple animals simultaneously and over extended periods, while automatically analyzing their motion. What may seem obvious marks a significant milestone, and paves the way for robust and accessible standardization and evaluation of such complex observations.
Imagine a researcher in the 19th century wearing a pith helmet, observing animals in their natural habitat. Or envision Konrad Lorenz, a veteran of the Max Planck Society, in the 1970s closely following his gray geese near Lake Starnberg—the beginning of behavioral research involved observing and recording what one sees.
The next step took place in the laboratory, where standardized environments were created to establish comparability. Researchers gained invaluable insights, but there were always limitations: the environment and test setup, the number of animals, and the duration of the observations did not correspond to the complexity of certain natural behaviors, either individual or social.
Moreover, observing animal behavior aims not only to better understand how specific species react to given stimuli, but also to help researchers better define mental disorders in humans in order to provide improved and individualized treatment.
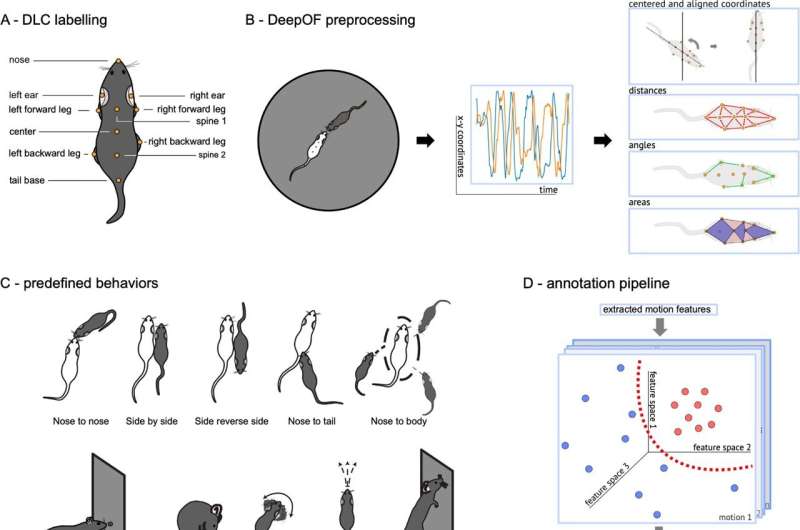
A few years ago, scientists achieved a breakthrough using the open-source toolbox DeepLabCut. They were not only able to track the center point of individual animals in simple environments but also automatically detect the complex body posture of multiple animals in real-world environments. This paved the way for the development of new tools capable of extracting information from these data, as capturing posture is not the same as analyzing the underlying behaviors.
Linking movement to behavior
Two research groups at the Max Planck Institute of Psychiatry took on this task. The teams led by Mathias V. Schmidt and Bertram Müller-Myhsok developed a Python package called DeepOF, which links the position of individual body markers over time with behavioral patterns. This allows them to analyze the behavior of animals, in their case mice, in a semi-natural environment in detail over any desired timeframe.
Two different approaches are used. In a supervised analysis pipeline, behaviors are predefined based on body postures over time, and the obtained data can be directly read and analyzed.
"Even more exciting is the unsupervised analysis pipeline," says statistician Müller-Myhsok. "Our program searches for similar behavioral episodes and classifies them," biologist Mathias Schmidt adds, "This approach opens up entirely new dimensions, enabling hypothesis-free automated investigation of complex social behavior and yielding highly interesting results."
This type of tool opens up new possibilities and brings behavioral biology, in terms of complexity, to a level comparable to molecular or functional biological analysis methods.
"In the future we can now better combine our results with other measurement dimensions, such as EEG recordings, neural activity data, or biosensor data," reports biologist Joeri Bordes. Lucas Miranda, the author of the DeepOF program, is enthusiastic about "open science" because "our program is freely available to researchers around the world, our code is of course open, and anyone is welcome to contribute to the project."
Nature Communications has given the program an independent seal of approval by publishing the teams' study. Moreover, thorough code and functionality reviews were assessed by the Journal of Open Source Software (JOSS). The program also represents an improvement for animal welfare, as the animals are subjected to fewer experiments.
Ultimately, the detailed analysis of behavior through this new dimension represents a significant step towards better translating the data regarding the exploration of human diseases and their treatment.
More information: Joeri Bordes et al, Automatically annotated motion tracking identifies a distinct social behavioral profile following chronic social defeat stress, Nature Communications (2023). DOI: 10.1038/s41467-023-40040-3
Journal information: Nature Communications
Provided by Max Planck Society