This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Scientists identify gene crucial for male meiosis during homologous pairing and synapsis
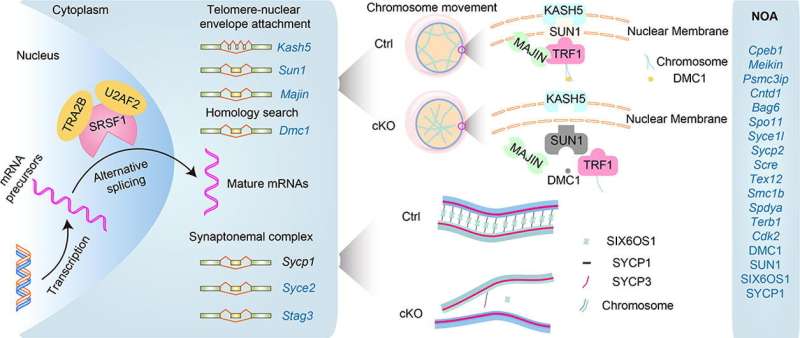
Meiotic recombination-related gene (e.g., DMC1, HFM1, MEIOB, MAJIN, C14ORF39/SIX6OS1, STAG3, SYCE1, SYCP2-3, TERB1-2) mutations have been identified in human subfertility or infertility. Surprisingly, most patients have been found to have aberrant splicing of genes such as MEIOB, C14ORF39/SIX6OS1, STAG3, and SYCE1. Therefore, it is imperative to understand the mechanism of alternative splicing (AS) and its role in human reproduction to provide new insights for clinical diagnosis.
It is well known that testes are rich in AS events. However, the underlying mechanisms of how AS functions in homologous pairing and synapsis are still largely unclear. A new study led by Dr. Jiali Liu (State Key Laboratory of Animal Biotech Breeding, College of Biological Sciences, China Agricultural University) and published in Science Bulletin shows that SRSF1 is crucial for male meiosis through alternative splicing during homologous pairing and synapsis in mice.
The team's previous research has shown that SRSF1 deficiency impairs primordial follicle formation and leads to primary ovarian insufficiency (POI). However, the underlying mechanisms by which SRSF1 regulates pre-mRNA splicing during homologous pairing and synapsis of meiotic prophase I in mouse spermatogenesis remain unknown.
This study reveals the critical role of an SRSF1-mediated post-transcriptional regulatory mechanism in homologous pairing and synapsis during meiotic prophase I, providing a framework for elucidating the molecular mechanisms underlying the post-transcriptional network of male meiosis.
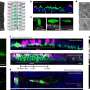
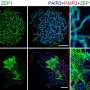
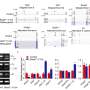
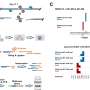
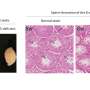
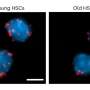
Conditional knockout of Srsf1 in mouse germ cells impaired homologous pairing and synapsis, leading to non-obstructive azoospermia (NOA). SRSF1 was required for initial homology recognition, telomere-led chromosome movement, and synaptonemal complex (SC) assembly. Moreover, SRSF1 interacted with TRA2B and U2AF2, directly binding and regulating the expression of Dmc1, Sycp1, Sun1, and Majin via AS to implement homologous pairing and synapsis during the meiotic prophase I program.
This study demonstrates that SRSF1-mediated post-transcriptional regulation is essential for homologous pairing and synapsis during the meiotic prophase I program. The discovery of the AS of NOA-related genes in a mouse model provides new insights for diagnosing human reproduction issues.
More information: Longjie Sun et al, SRSF1 is crucial for male meiosis through alternative splicing during homologous pairing and synapsis in mice, Science Bulletin (2023). DOI: 10.1016/j.scib.2023.04.030
Provided by Science China Press