This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Promoting microbiome data standards to advance research
Researchers have predicted that our planet may be home to 1 trillion species of microbes. Communities of these microorganisms, called microbiomes that exist in particular environments, are incredibly important to the health of everything from individual people to complete ecosystems. To study microbiomes, scientists need to collect, process, and share data in a standardized way. Researchers at the National Microbiome Data Collaborative (NMDC) are seeking to develop community-supported microbiome data standards.
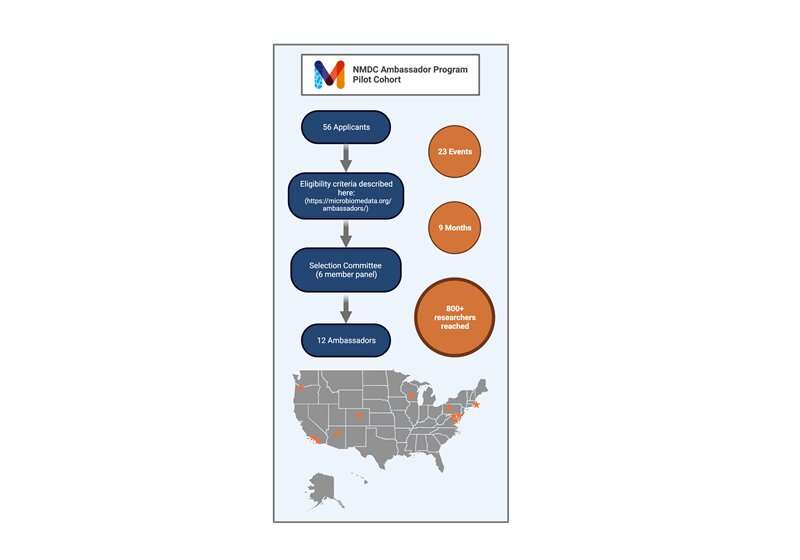
In a recently published Nature Microbiology article, researchers look at the impacts of a community learning Ambassador Program. The Ambassador Program was launched by the NMDC in 2021 to encourage awareness and adoption of microbiome data and metadata standards among the research community engaged in the collection and analysis of microbiome data. During the Ambassador program's year-long term, more than 800 researchers attended 23 Ambassador-hosted presentations.
From 2021 to 2022, NMDC team members trained their community-learning cohort about the importance of metadata standards. The Ambassadors then used the training materials to host their own events and workshops. These events helped to spread the NMDC mission and broadened the NMDC community network by reaching more diverse audiences. The program supported workforce development by honing the Ambassadors' presentation skills. It also provided key networking opportunities at a wide range of venues.
Researchers have produced an incredible amount of data about the microbes that exist across many environments. The information that is collected in conjunction with this data is called metadata. For example, metadata could include information about where a particular sample was found. Once researchers have gathered data and metadata, they want to share them with others. But unless data are collected, processed, and shared in a standardized way, other researchers have difficulties reusing them.
The NMDC launched the Ambassador Program to educate researchers about the benefits of metadata standards. Out of 56 applicants, the program accepted 12 early-career researchers from universities, government agencies, and research organizations. The NMDC team hosted six training sessions for the Ambassadors. These sessions included best practices in data management, data standards, and community engagement. The Ambassadors then hosted in-person and virtual events with their respective microbiome research communities.
Insights from the cohort provided the NMDC team with ideas for how to improve future iterations of the program. The NMDC team recruits early-career researchers on an annual basis to join the Ambassadors Program. The 2023 cohort of the NMDC Ambassador Program has an expanded scope that focuses on multiple aspects of microbiome data stewardship, including methods for data collection, processing, and reuse. Over time, the Ambassador Program aims to promote better use and reuse of microbiome data.
More information: Julia M. Kelliher et al, Cohort-based learning for microbiome research community standards, Nature Microbiology (2023). DOI: 10.1038/s41564-023-01361-7
Journal information: Nature Microbiology
Provided by US Department of Energy